4WTZ
 
 | | Human CEACAM6-CEACAM8 N-domain heterodimer complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 6, Carcinoembryonic antigen-related cell adhesion molecule 8, NICKEL (II) ION | | Authors: | Kirouac, K.N, Prive, G.G. | | Deposit date: | 2014-10-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Human CEACAM6-CEACAM8 N-domain heterodimer complex
To Be Published
|
|
2EDP
 
 | |
2E2N
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in the apo form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEXOKINASE, SULFATE ION | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
5OB1
 
 | |
2E2O
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with glucose | | Descriptor: | HEXOKINASE, beta-D-glucopyranose | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
6APM
 
 | | Hen egg-white lysozyme (WT), solved with serial millisecond crystallography using synchrotron radiation | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Lyubimov, A.Y, Mathews, I.I, Uervivojnangkoorn, M, Soltis, S.M, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
2ED0
 
 | | Solution structure of the SH3 domain of Abl interactor 2 (Abelson interactor 2) | | Descriptor: | Abl interactor 2 | | Authors: | Abe, H, Tochio, N, Miyamoto, K, Saito, K, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of Abl interactor 2 (Abelson interactor 2)
To be Published
|
|
2C8E
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (Free state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8B
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form II) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
5OB0
 
 | |
2CI8
 
 | | sh2 domain of human nck1 adaptor protein - uncomplexed | | Descriptor: | CYTOPLASMIC PROTEIN NCK1, PENTAETHYLENE GLYCOL, SULFATE ION | | Authors: | Frese, S, Schubert, W.-D, Findeis, A.C, Marquardt, T, Roske, Y.S, Stradal, T.E.B, Heinz, D.W. | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
J.Biol.Chem., 281, 2006
|
|
2CG7
 
 | | SECOND AND THIRD FIBRONECTIN TYPE I MODULE PAIR (CRYSTAL FORM II). | | Descriptor: | FIBRONECTIN | | Authors: | Rudino-Pinera, E, Ravelli, R.B.G, Sheldrick, G.M, Nanao, M.H, Werner, J.M, Schwarz-Linek, U, Potts, J.R, Garman, E.F. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Solution and Crystal Structures of a Module Pair from the Staphylococcus Aureus-Binding Site of Human Fibronectin-A Tale with a Twist.
J.Mol.Biol., 368, 2007
|
|
2C89
 
 | | Structure of the wild-type C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2CI9
 
 | | Nck1 SH2-domain in complex with a dodecaphosphopeptide from EPEC protein Tir | | Descriptor: | CYTOPLASMIC PROTEIN NCK1, TRANSLOCATED INTIMIN RECEPTOR | | Authors: | Frese, S, Schubert, W.-D, Findeis, A.C, Marquardt, T, Roske, Y.S, Stradal, T.E.B, Heinz, D.W. | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
J.Biol.Chem., 281, 2006
|
|
2J5U
 
 | | MreC Lysteria monocytogenes | | Descriptor: | MREC PROTEIN | | Authors: | van den Ent, F, Leaver, M, Bendezu, F, Errington, J, de Boer, P, Lowe, J. | | Deposit date: | 2006-09-19 | | Release date: | 2006-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric Structure of the Cell Shape Protein Mrec and its Functional Implications.
Mol.Microbiol., 62, 2006
|
|
2D8J
 
 | | Solution structure of the SH3 domain of Fyn-related kinase | | Descriptor: | fyn-related kinase | | Authors: | Qin, X.R, Kurosaki, C, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of Fyn-related kinase
TO BE PUBLISHED
|
|
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
5OAV
 
 | |
8BVW
 
 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
2FV8
 
 | | The crystal structure of RhoB in the GDP-bound state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Rho-related GTP-binding protein RhoB | | Authors: | Turnbull, A.P, Soundararajan, M, Smee, C, Johansson, C, Schoch, G, Gorrec, F, Bray, J, Papagrigoriou, E, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of RhoB in the GDP-bound state
To be Published
|
|
2JM0
 
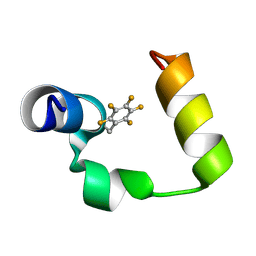 | | Solution structure of chicken villin headpiece subdomain containing a fluorinated side chain in the core | | Descriptor: | Villin-1 | | Authors: | Cornilescu, C.C, Cornilescu, G, Hadley, E.B, Gellman, S.H, Markley, J.L. | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a small protein containing a fluorinated side chain in the core.
Protein Sci., 16, 2007
|
|
2JT3
 
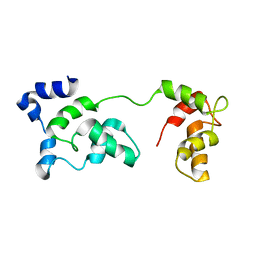 | | Solution Structure of F153W cardiac troponin C | | Descriptor: | Troponin C | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
6ZON
 
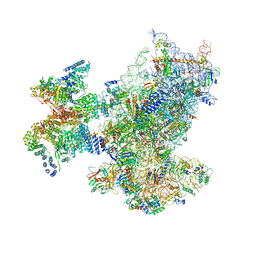 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 1 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
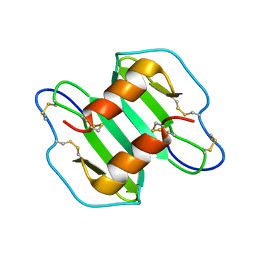
2K01
 
 | |
2K05
 
 | |
