2OEW
 
 | | Structure of ALIX/AIP1 Bro1 Domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
3TAV
 
 | |
4E2H
 
 | |
4E3L
 
 | |
4E2S
 
 | |
3TCR
 
 | |
4DX5
 
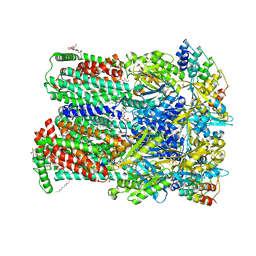 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, Acriflavine resistance protein B, DARPIN, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2NSC
 
 | |
3G20
 
 | | Crystal structure of the major pseudopilin from the type 2 secretion system of enterohaemorrhagic Escherichia coli | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Korotkov, K.V, Gray, M.D, Kreger, A, Turley, S, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2009-01-30 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Calcium is essential for the major pseudopilin in the type 2 secretion system.
J.Biol.Chem., 284, 2009
|
|
3G2O
 
 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with (S)-adenosyl-L-methionine (SAM) | | Descriptor: | PCZA361.24, S-ADENOSYLMETHIONINE | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
2H3R
 
 | | Crystal structure of ORF52 from Murid herpesvirus 4 (MuHV-4) (Murine gammaherpesvirus 68). Northeast Structural Genomics Consortium target MhR28B. | | Descriptor: | Hypothetical protein BQLF2 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Ho, C.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-23 | | Release date: | 2006-08-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of ORF52 from Murid herpesvirus 4 (MuHV-4) (Murine gammaherpesvirus 68). Northeast Structural Genomics Consortium target MhR28B.
To be Published
|
|
3RM3
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
4ILK
 
 | | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MANGANESE (II) ION, Starvation sensing protein rspB, ... | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-12-31 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH
To be Published
|
|
2OIP
 
 | |
2NVH
 
 | |
3RQI
 
 | |
3R9J
 
 | | 4.3A resolution structure of a MinD-MinE(I24N) protein complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
4IBT
 
 | | Human p53 core domain with hot spot mutation R273H and second-site suppressor mutation T284R | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Diskin-Posner, Y, Eldar, A, Rozenberg, H, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
4IE4
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with 5-carboxy-8-hydroxyquinoline (IOX1) | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5048 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
3RSI
 
 | |
2O02
 
 | | Phosphorylation independent interactions between 14-3-3 and Exoenzyme S: from structure to pathogenesis | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, ExoS (416-430) peptide | | Authors: | Ottmann, C, Yasmin, L, Weyand, M, Hauser, A.R, Wittinghofer, A, Hallberg, B. | | Deposit date: | 2006-11-27 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphorylation-independent interaction between 14-3-3 and exoenzyme S: from structure to pathogenesis
Embo J., 26, 2007
|
|
2NZ8
 
 | | N-terminal DHPH cassette of Trio in complex with nucleotide-free Rac1 | | Descriptor: | ras-related C3 botulinum toxin substrate 1 isoform Rac1, triple functional domain protein | | Authors: | Chhatriwala, M.K, Betts, L, Worthylake, D.K, Sondek, J. | | Deposit date: | 2006-11-22 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The DH and PH Domains of Trio Coordinately Engage Rho GTPases for their Efficient Activation
J.Mol.Biol., 368, 2007
|
|
4EBU
 
 | | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-24 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I
TO BE PUBLISHED
|
|
3RCD
 
 | | HER2 Kinase Domain Complexed with TAK-285 | | Descriptor: | N-{2-[4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethyl}-3-hydroxy-3-methylbutanamide, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Aertgeerts, K, Skene, R, Sogabe, S. | | Deposit date: | 2011-03-30 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Design and Synthesis of Novel Human Epidermal Growth Factor Receptor 2 (HER2)/Epidermal Growth Factor Receptor (EGFR) Dual Inhibitors Bearing a Pyrrolo[3,2-d]pyrimidine Scaffold.
J.Med.Chem., 54, 2011
|
|
3DMU
 
 | | Crystal structure of Staphylococcal nuclease variant PHS T62K at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Thermonuclease | | Authors: | Khangulov, V.S, Schlessman, J.L, Garcia-Moreno, E.B, Benning, M, Isom, D. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant PHS T62K at cryogenic temperature
To be Published
|
|
