2YNZ
 
 | |
2YO3
 
 | |
6PDA
 
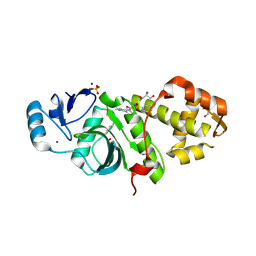 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 74 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-(pyridin-2-yl)benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6PDE
 
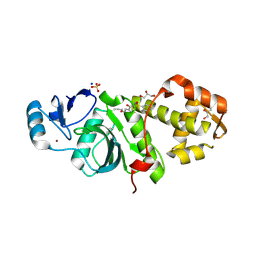 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 40 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-propoxybenzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
4KVZ
 
 | |
7PP9
 
 | |
4XG1
 
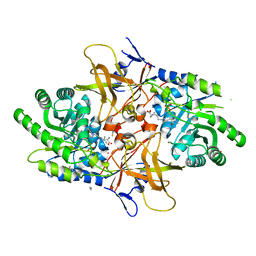 | | Psychromonas ingrahamii diaminopimelate decarboxylase with LLP | | Descriptor: | (2S)-2-amino-6-[[3-hydroxy-2-methyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]hexanoic acid, Diaminopimelate decarboxylase, POTASSIUM ION, ... | | Authors: | Peverelli, M.G, Wubben, J.M, Panjikar, S, Perugini, M.A. | | Deposit date: | 2014-12-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Expression to crystallization of diaminopimelate decarboxylase from the psychrophile Psychromonas ingrahamii
To Be Published
|
|
4XJS
 
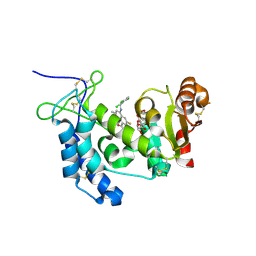 | | Human CD38 complexed with inhibitor 1 [6-fluoro-2-methyl-4-[(2,3,6-trichlorobenzyl)amino]quinoline-8-carboxamide] | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, 6-fluoro-2-methyl-4-[(2,3,6-trichlorobenzyl)amino]quinoline-8-carboxamide, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 | | Authors: | Shewchuk, L.M, Deaton, D, Stewart, E. | | Deposit date: | 2015-01-09 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 4-Amino-8-quinoline Carboxamides as Novel, Submicromolar Inhibitors of NAD-Hydrolyzing Enzyme CD38.
J.Med.Chem., 58, 2015
|
|
4LDT
 
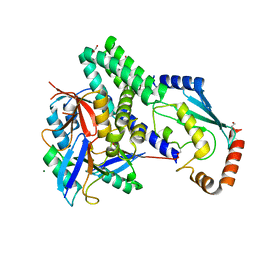 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBCH5B~Ub | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin, ... | | Authors: | Wiener, R, DiBello, A.T, Lombardi, P.M, Guzzo, C.M, Zhang, X, Matunis, M.J, Wolberger, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1YTT
 
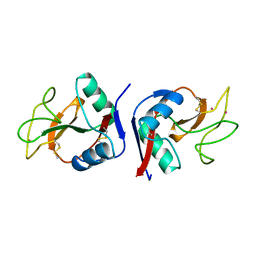 | | YB SUBSTITUTED SUBTILISIN FRAGMENT OF MANNOSE BINDING PROTEIN-A (SUB-MBP-A), MAD STRUCTURE AT 110K | | Descriptor: | MANNOSE-BINDING PROTEIN A, YTTERBIUM (III) ION | | Authors: | Burling, F.T, Weis, W.I, Flaherty, K.M, Brunger, A.T. | | Deposit date: | 1995-11-09 | | Release date: | 1996-06-10 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of protein solvation and discrete disorder with experimental crystallographic phases.
Science, 271, 1996
|
|
8R3V
 
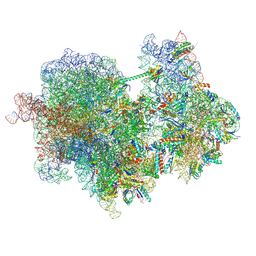 | |
7B2G
 
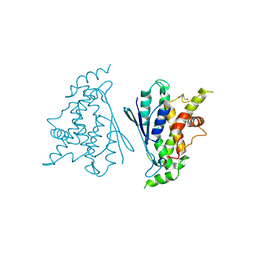 | | Crystal structure of R120Q GDAP1 mutant | | Descriptor: | GLYCEROL, Ganglioside-induced differentiation-associated protein 1 | | Authors: | Nguyen, G.T.T, Sutinen, A, Kursula, P. | | Deposit date: | 2020-11-26 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conserved intramolecular networks in GDAP1 are closely connected to CMT-linked mutations and protein stability.
Plos One, 18, 2023
|
|
175L
 
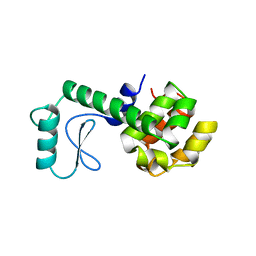 | |
1YJC
 
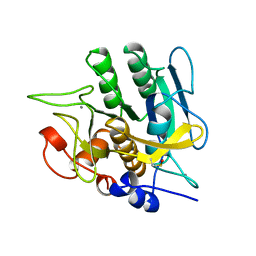 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 50% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YJA
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 20% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1AL2
 
 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT V1160I | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-06-09 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
3BFB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the 9-keto-2(E)-decenoic acid | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
4XJT
 
 | | Human CD38 complexed with inhibitor 2 [4-[(2,6-dimethylbenzyl)amino]-2-methylquinoline-8-carboxamide] | | Descriptor: | 4-[(2,6-dimethylbenzyl)amino]-2-methylquinoline-8-carboxamide, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Shewchuk, L.M, Deaton, D.N, Stewart, E. | | Deposit date: | 2015-01-09 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 4-Amino-8-quinoline Carboxamides as Novel, Submicromolar Inhibitors of NAD-Hydrolyzing Enzyme CD38.
J.Med.Chem., 58, 2015
|
|
4MRW
 
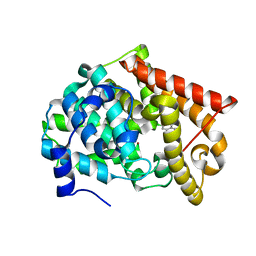 | | Crystal structure of PDE10A2 with fragment ZT0120 (7-chloroquinolin-4-ol) | | Descriptor: | 7-chloroquinolin-4-ol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, NIenaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
169L
 
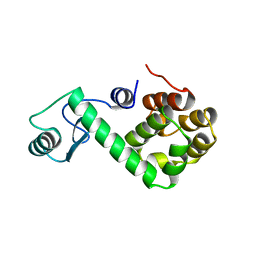 | |
168L
 
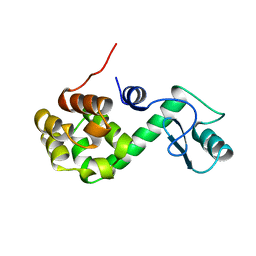 | |
6BG8
 
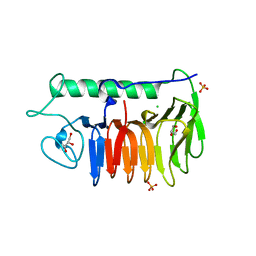 | |
6PD9
 
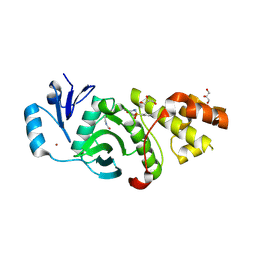 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 60 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-(1H-pyrazol-1-yl)benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6SJC
 
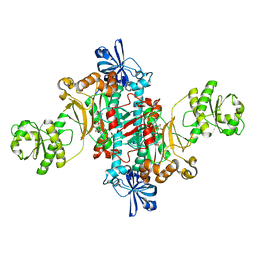 | | Structure of T. thermophilus AspRS in Complex with 5'-O-(N-(L-aspartyl)-sulfamoyl)adenosine | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, Aspartate--tRNA(Asp) ligase | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
8W9Z
 
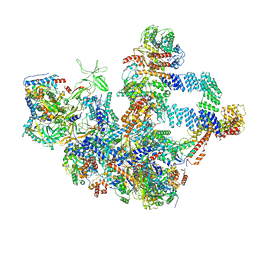 | | The cryo-EM structure of the Nicotiana tabacum PEP-PAP | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta'', ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
