3ZG8
 
 | | Crystal Structure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZG9
 
 | |
3ZGA
 
 | | Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN 4 | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZGB
 
 | | Greater efficiency of photosynthetic carbon fixation due to single amino acid substitution | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, PHOSPHOENOLPYRUVATE CARBOXYLASE, ... | | Authors: | Paulus, J.K, Schlieper, D, Groth, G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Greater Efficiency of Photosynthetic Carbon Fixation due to Single Amino Acid Substitution
Nat.Commun., 4, 2013
|
|
3ZGC
 
 | | crystal structure of the KEAP1-NEH2 complex | | Descriptor: | ACETATE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1, NUCLEAR FACTOR ERYTHROID 2-RELATED FACTOR 2 | | Authors: | Hoerer, S, Reinert, D, Ostmann, K, Hoevels, Y, Nar, H. | | Deposit date: | 2012-12-17 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal-Contact Engineering to Obtain a Crystal Form of the Kelch Domain of Human Keap1 Suitable for Ligand-Soaking Experiments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3ZGD
 
 | | crystal structure of a KEAP1 mutant | | Descriptor: | ACETATE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1, SODIUM ION | | Authors: | Hoerer, S, Reinert, D, Ostmann, K, Hoevels, Y, Nar, H. | | Deposit date: | 2012-12-17 | | Release date: | 2013-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal-Contact Engineering to Obtain a Crystal Form of the Kelch Domain of Human Keap1 Suitable for Ligand-Soaking Experiments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3ZGE
 
 | | Greater efficiency of photosynthetic carbon fixation due to single amino acid substitution | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, C4 PHOSPHOENOLPYRUVATE CARBOXYLASE, ... | | Authors: | Paulus, J.K, Schlieper, D, Groth, G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Greater Efficiency of Photosynthetic Carbon Fixation due to Single Amino Acid Substitution
Nat.Commun., 4, 2013
|
|
3ZGF
 
 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with in complex with NPE caged UDP-Gal (P2(1)2(1)2(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3ZGG
 
 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with NPE caged UDP-Gal (C222(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, GLYCEROL, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3ZGH
 
 | | Crystal structure of the KRT10-binding region domain of the pneumococcal serine rich repeat protein PsrP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CELL WALL SURFACE ANCHOR FAMILY PROTEIN, ... | | Authors: | Schulte, T, Loefling, J, Mikaelsson, C, Kikhney, A, Hentrich, K, Diamante, A, Ebel, C, Normark, S, Svergun, D, Henriques-Normark, B, Achour, A. | | Deposit date: | 2012-12-17 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Basic Keratin 10-Binding Domain of the Virulence-Associated Pneumococcal Serine-Rich Protein Psrp Adopts a Novel Mscramm Fold.
Open Biol., 4, 2014
|
|
3ZGI
 
 | | Crystal structure of the KRT10-binding region domain of the pneumococcal serine rich repeat protein PsrP | | Descriptor: | 1,2-ETHANEDIOL, CELL WALL SURFACE ANCHOR FAMILY PROTEIN, SULFATE ION | | Authors: | Schulte, T, Loefling, J, Mikaelsson, C, Kikhney, A, Hentrich, K, Diamante, A, Ebel, C, Normark, S, Svergun, D, Henriques-Normark, B, Achour, A. | | Deposit date: | 2012-12-17 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Basic Keratin 10-Binding Domain of the Virulence-Associated Pneumococcal Serine-Rich Protein Psrp Adopts a Novel Mscramm Fold.
Open Biol., 4, 2014
|
|
3ZGJ
 
 | |
3ZGK
 
 | | NMR solution structure of the RXLR effector AVR3a11 from Phytophthora Capsici | | Descriptor: | AVR3A11 | | Authors: | Tolchard, J, Chambers, V.S, Boutemy, L.S, Gathercole, R.L, Banfield, M.J, Blumenschein, T.M. | | Deposit date: | 2012-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Avr3A11 from Phytophthora Capsi
To be Published
|
|
3ZGL
 
 | | Crystal structures of Escherichia coli IspH in complex with AMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-4-amino-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
3ZGN
 
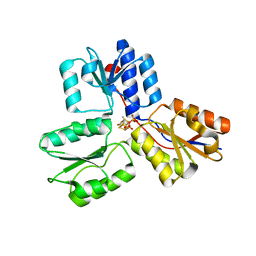 | | Crystal structures of Escherichia coli IspH in complex with TMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-3-methyl-4-sulfanylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
3ZGO
 
 | |
3ZGP
 
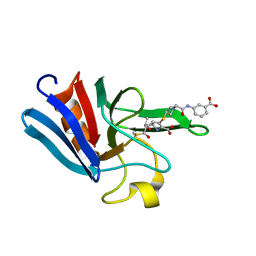 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase acylated by ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Triboulet, S, Dubee, V, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-18 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3ZGQ
 
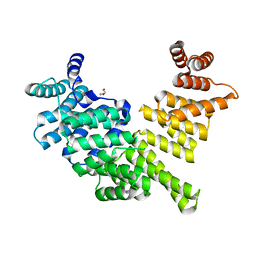 | | Crystal structure of human interferon-induced protein IFIT5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, INTERFERON-INDUCED PROTEIN WITH TETRATRICOPEPTIDE REPEATS 5 | | Authors: | Katibah, G.E, Lee, H.J, Huizar, J.P, Vogan, J.M, Alber, T, Collins, K. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | TRNA Binding, Structure, and Localization of the Human Interferon-Induced Protein Ifit5.
Mol.Cell, 49, 2013
|
|
3ZGV
 
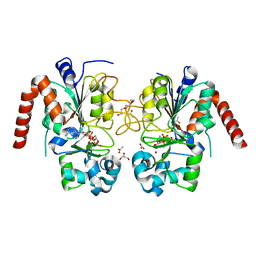 | | Structure of human SIRT2 in complex with ADP-ribose | | Descriptor: | ACETATE ION, GLYCEROL, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, ... | | Authors: | Moniot, S, Steegborn, C. | | Deposit date: | 2012-12-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure Analysis of Human Sirt2 and its Adp-Ribose Complex
J.Struct.Biol., 182, 2013
|
|
3ZGX
 
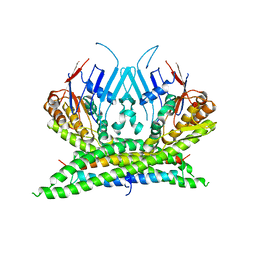 | | Crystal structure of the kleisin-N SMC interface in prokaryotic condensin | | Descriptor: | CHROMOSOME PARTITION PROTEIN SMC, SEGREGATION AND CONDENSATION PROTEIN A | | Authors: | Burmann, F, Shin, H, Basquin, J, Soh, Y, Gimenez, V, Kim, Y, Oh, B, Gruber, S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Asymmetric Smc-Kleisin Bridge in Prokaryotic Condensin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3ZGY
 
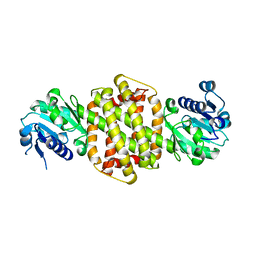 | |
3ZGZ
 
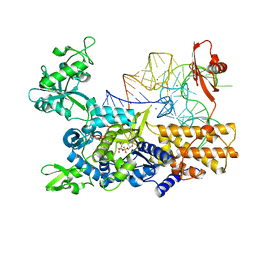 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and toxic moiety from agrocin 84 (TM84) in aminoacylation-like conformation | | Descriptor: | LEUCINE--TRNA LIGASE, MAGNESIUM ION, TRNA-LEU UAA ISOACCEPTOR, ... | | Authors: | Chopra, S, Palencia, A, Virus, C, Tripathy, A, Temple, B.R, Velazquez-Campoy, A, Cusack, S, Reader, J.S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Plant Tumour Biocontrol Agent Employs a tRNA-Dependent Mechanism to Inhibit Leucyl-tRNA Synthetase
Nat.Commun., 4, 2013
|
|
3ZH0
 
 | | Functional and structural role of the N-terminal extension in Methanosarcina acetivorans protoglobin | | Descriptor: | FORMIC ACID, GLYCEROL, PROTOGLOBIN, ... | | Authors: | Ciaccio, C, Pesce, A, Tundo, G.R, Tilleman, L, Dewilde, S, Moens, L, Ascenzi, P, Bolognesi, M, Nardini, M, Coletta, M. | | Deposit date: | 2012-12-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Role of the N-Terminal Extension in Methanosarcina Acetivorans Protoglobin.
Biochim.Biophys.Acta, 1834, 2013
|
|
3ZH2
 
 | | Structure of Plasmodium falciparum lactate dehydrogenase in complex with a DNA aptamer | | Descriptor: | DNA APTAMER, L-LACTATE DEHYDROGENASE | | Authors: | Cheung, Y.W, Kwok, J, Law, A.W.L, Watt, R.M, Kotaka, M, Tanner, J.A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminatory Recognition of Plasmodium Lactate Dehydrogenase by a DNA Aptamer
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZH3
 
 | |
