5J1H
 
 | |
5IY7
 
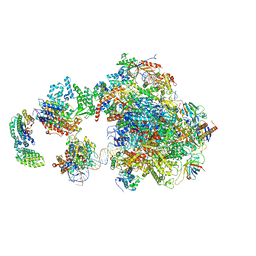 | | Human holo-PIC in the open state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
7A5G
 
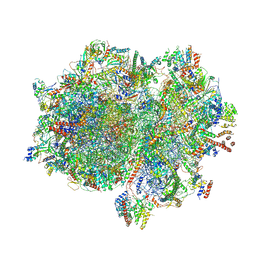 | | Structure of the elongating human mitoribosome bound to mtEF-Tu.GMPPCP and A/T mt-tRNA | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
5IZ1
 
 | | Physcomitrella patens FBPase | | Descriptor: | fructose-1,6-bisphosphatase | | Authors: | Einsle, O, Guetle, D. | | Deposit date: | 2016-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7A7D
 
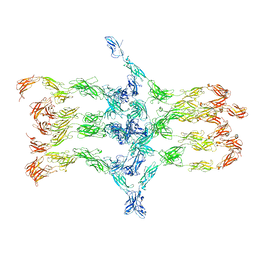 | | Cadherin fit into cryo-ET map | | Descriptor: | Desmocollin-2, Desmoglein-2 | | Authors: | Sikora, M, Ermel, U.H, Seybold, A, Kunz, M, Calloni, G, Reitz, J, Vabulas, R.M, Hummer, G, Frangakis, A.S. | | Deposit date: | 2020-08-28 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Desmosome architecture derived from molecular dynamics simulations and cryo-electron tomography.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5J1I
 
 | |
3O95
 
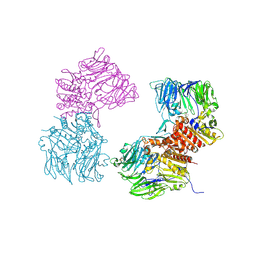 | | Crystal Structure of Human DPP4 Bound to TAK-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a 3-Pyridylacetic Acid Derivative (TAK-100) as a Potent, Selective and Orally Active Dipeptidyl Peptidase IV (DPP-4) Inhibitor.
J.Med.Chem., 53, 2011
|
|
7A5K
 
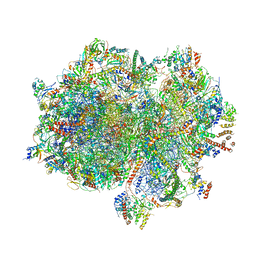 | | Structure of the human mitoribosome in the post translocation state bound to mtEF-G1 | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
6NJM
 
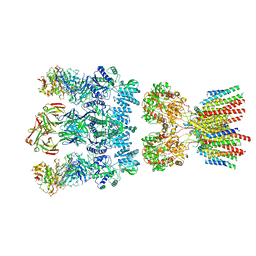 | | Architecture and subunit arrangement of native AMPA receptors | | Descriptor: | 15F1 Fab heavy chain, 15F1 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Zhao, Y. | | Deposit date: | 2019-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM.
Science, 364, 2019
|
|
5EHM
 
 | |
6ZTO
 
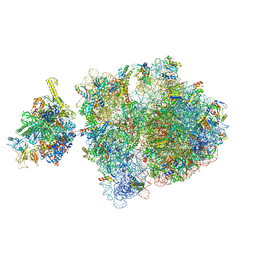 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
7A5I
 
 | | Structure of the human mitoribosome with A- P-and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7U34
 
 | | The structure of phosphoglucose isomerase from Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Yan, K, Kowalski, B, Fang, W, van Aalten, D. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
5J1G
 
 | |
6ZM5
 
 | | Human mitochondrial ribosome in complex with OXA1L, mRNA, A/A tRNA, P/P tRNA and nascent polypeptide | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Andrell, J, Amunts, A. | | Deposit date: | 2020-07-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanism of membrane-tethered mitochondrial protein synthesis.
Science, 371, 2021
|
|
5J3J
 
 | | Crystal Structure of human DPP-IV in complex with HL1 | | Descriptor: | (2~{S},3~{R})-8,9-dimethoxy-3-[2,4,5-tris(fluoranyl)phenyl]-2,3-dihydro-1~{H}-benzo[f]chromen-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Wu, F, Li, H, Zhao, Z, Zhu, L, Xu, H, Li, S. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of human DPP-IV in complex with HL1
To Be Published
|
|
6ZNS
 
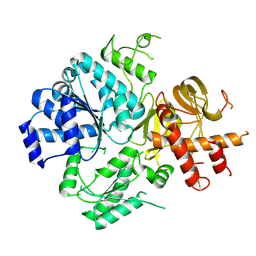 | | Crystal Structure of DUF1998 helicase MrfA | | Descriptor: | Uncharacterized ATP-dependent helicase YprA, ZINC ION | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
5EQG
 
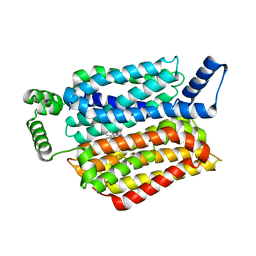 | | Human GLUT1 in complex with inhibitor (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide | | Descriptor: | (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6ZSE
 
 | | Human mitochondrial ribosome in complex with mRNA, A/P-tRNA and P/E-tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
5J5B
 
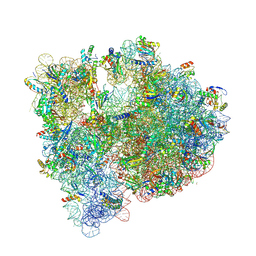 | | Structure of the WT E coli ribosome bound to tetracycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-27 | | Last modified: | 2018-08-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2022-02-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5DLJ
 
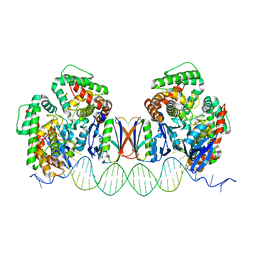 | | Crystal Structure of Cas-DNA-N1 complex | | Descriptor: | 39-mer DNA N1-F, 39-mer DNA N1-R, CRISPR-associated endonuclease Cas1, ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-05 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
6NTM
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-232 in Complex with the Reactivator, HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
3O9V
 
 | | Crystal Structure of Human DPP4 Bound to TAK-986 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(aminomethyl)-2-methyl-4-(4-methylphenyl)-6-(2-methylpropyl)pyridine-3-carboxic acid, ... | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2010-08-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of a 3-Pyridylacetic Acid Derivative (TAK-100) as a Potent, Selective and Orally Active Dipeptidyl Peptidase IV (DPP-4) Inhibitor.
J.Med.Chem., 53, 2011
|
|
5DQZ
 
 | | Crystal Structure of Cas-DNA-PAM complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (36-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
