6W4O
 
 | | CaMKII alpha-30 Cryo-EM reconstruction | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Chao, L.H, Stratton, M.M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub-dependent regulation.
Sci.Signal., 13, 2020
|
|
2LHB
 
 | |
6W4P
 
 | | CaMKII alpha-30 Cryo-EM reconstruction - Class B | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Chao, L.H, Stratton, M.M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub-dependent regulation.
Sci.Signal., 13, 2020
|
|
8K1N
 
 | | mycobacterial efflux pump, substrate-bound state | | Descriptor: | CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, Multidrug efflux system permease protein Rv1217c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6C04
 
 | | Mtb RNAP Holo/RbpA/double fork DNA -closed clamp | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
8P7Y
 
 | | Mycoplasma pneumoniae 70S ribosome with second S4 protein on large subunit | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Schacherl, M, Xue, L, Spahn, C.M.T, Mahamid, J. | | Deposit date: | 2023-05-31 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7A8P
 
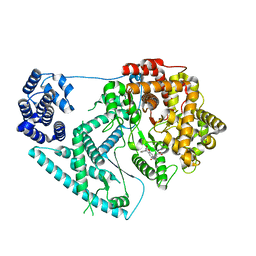 | | Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor. | | Descriptor: | (3~{R})-1-[(2~{R})-2-[4-(2-chloranyl-4-fluoranyl-phenyl)-2-oxidanylidene-chromen-7-yl]oxypropanoyl]piperidine-3-carboxylic acid, DNA-directed RNA polymerase, mitochondrial | | Authors: | Hillen, H.S, Bonekamp, N, Peter, B, Felser, A, Bergbrede, T, Choidas, A, Horn, M, Unger, A, di Lucrezia, R, Atanassov, I, Li, X, Koch, U, Menninger, S, Boros, J, Habenberger, P, Giavalisco, P, Cramer, P, Denzel, M, Nussbaumer, P, Klebl, B, Falkenberg, M, Gustafsson, C.M, Larsson, N.G. | | Deposit date: | 2020-08-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Small-molecule inhibitors of human mitochondrial DNA transcription.
Nature, 588, 2020
|
|
8UUZ
 
 | | Campylobacter jejuni CosR apo form | | Descriptor: | DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
2M0M
 
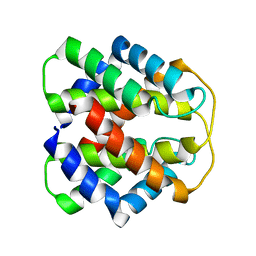 | | Structural Characterization of Minor Ampullate Spidroin Domains and their Distinct Roles in Fibroin Solubility and Fiber Formation | | Descriptor: | Minor ampullate fibroin 1 | | Authors: | Yang, D, Gao, Z, Lin, Z, Huang, W, Lai, C, Fan, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of minor ampullate spidroin domains and their distinct roles in fibroin solubility and fiber formation
Plos One, 8, 2013
|
|
2MIX
 
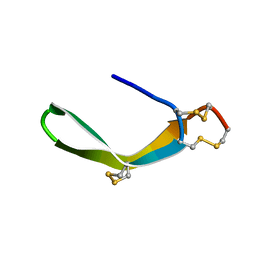 | | Structure of a novel venom peptide toxin from sample limited terebrid marine snail | | Descriptor: | venom peptide toxin | | Authors: | Bhuiyan, M.H, Anand, P, Grigoryan, A, Holford, M, Poget, S.F. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Sample limited characterization of a novel disulfide-rich venom peptide toxin from terebrid marine snail Terebra variegata.
Plos One, 9, 2014
|
|
8GIZ
 
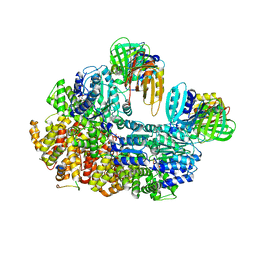 | | E. coli clamp loader with open clamp | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GIY
 
 | | E. coli clamp loader with closed clamp | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8WZZ
 
 | |
1J3H
 
 | | Crystal structure of apoenzyme cAMP-dependent protein kinase catalytic subunit | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Wu, J, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic Features of cAMP-dependent Protein Kinase Revealed by Apoenzyme Crystal Structure
J.Mol.Biol., 327, 2003
|
|
6XW2
 
 | | Crystal structure of the bright genetically encoded calcium indicator NCaMP7 based on mNeonGreen fluorescent protein | | Descriptor: | CALCIUM ION, Genetically encoded calcium indicator NCaMP7 based on mNeonGreen fluorescent protein, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Lazarenko, V.A, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Genetically Encoded Bright Positive Calcium Indicator NCaMP7 Based on the mNeonGreen Fluorescent Protein.
Int J Mol Sci, 21, 2020
|
|
7YQQ
 
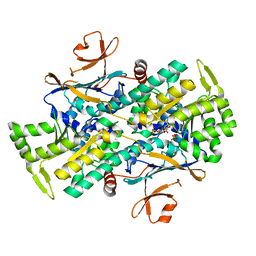 | | Crystal Structure of Xcc NAMPT and its complex with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, PHOSPHATE ION, Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
7YQR
 
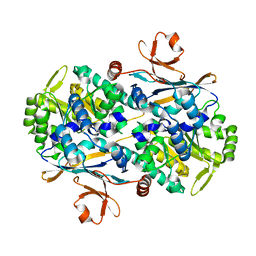 | | Crystal Structure of Xcc NAMPT and its complex with NAM | | Descriptor: | NICOTINAMIDE, Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
1H8S
 
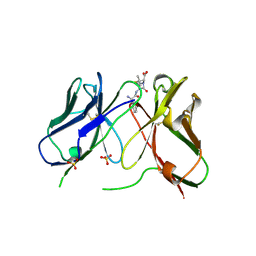 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment complexed with the hapten. | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-15 | | Release date: | 2001-08-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
3L63
 
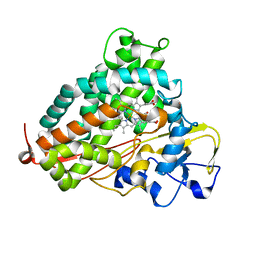 | | Crystal structure of camphor-bound P450cam at low [K+] | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Rupniewski, I, Goodin, D.B. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450cam visits an open conformation in the absence of substrate.
Biochemistry, 49, 2010
|
|
1CMK
 
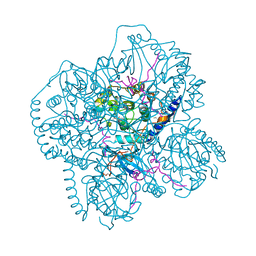 | | CRYSTAL STRUCTURES OF THE MYRISTYLATED CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE REVEAL OPEN AND CLOSED CONFORMATIONS | | Descriptor: | IODIDE ION, MYRISTIC ACID, cAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT, ... | | Authors: | Zheng, J, Knighton, D.R, Xuong, N.-H, Taylor, S.S, Sowadski, J.M, Ten Eyck, L.F. | | Deposit date: | 1993-11-18 | | Release date: | 1994-05-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations.
Protein Sci., 2, 1993
|
|
6WFL
 
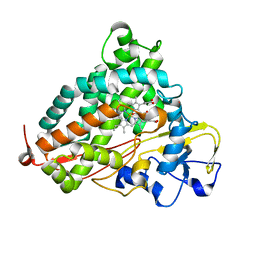 | | Camphor soaked P450cam D251E | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Amaya, J.A, Poulos, T.L, Batabyal, D. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proton Relay Network in the Bacterial P450s: CYP101A1 and CYP101D1.
Biochemistry, 59, 2020
|
|
1CTP
 
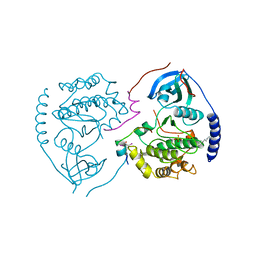 | | STRUCTURE OF THE MAMMALIAN CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND AN INHIBITOR PEPTIDE DISPLAYS AN OPEN CONFORMATION | | Descriptor: | MYRISTIC ACID, cAMP-DEPENDENT PROTEIN KINASE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Karlsson, R, Zheng, J, Xuong, N.H, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 1993-04-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the mammalian catalytic subunit of cAMP-dependent protein kinase and an inhibitor peptide displays an open conformation.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
5AEC
 
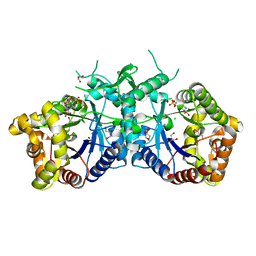 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1FMO
 
 | | CRYSTAL STRUCTURE OF A POLYHISTIDINE-TAGGED RECOMBINANT CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH THE PEPTIDE INHIBITOR PKI(5-24) AND ADENOSINE | | Descriptor: | ADENOSINE, CAMP-DEPENDENT PROTEIN KINASE, HEAT STABLE RABBIT SKELETAL MUSCLE INHIBITOR PROTEIN | | Authors: | Narayana, N, Cox, S, Shaltiel, S, Taylor, S.S, Xuong, N.-H. | | Deposit date: | 1997-07-08 | | Release date: | 1998-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a polyhistidine-tagged recombinant catalytic subunit of cAMP-dependent protein kinase complexed with the peptide inhibitor PKI(5-24) and adenosine.
Biochemistry, 36, 1997
|
|
1XHY
 
 | | X-ray structure of the Y702F mutant of the GluR2 ligand-binding core (S1S2J) in complex with kainate at 1.85 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor, SULFATE ION | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
