6NFF
 
 | | Structure of X-prolyl dipeptidyl aminopeptidase from Lactobacillus helveticus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Juers, D.H, Bratt, N.J, Ojennus, D.D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a prolyl aminodipeptidase (PepX) from Lactobacillus helveticus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6H36
 
 | | The crystal structure of human carbonic anhydrase VII in complex with 4-(4-phenylpiperidine-1-carbonyl)benzenesulfonamide. | | Descriptor: | 4-(4-phenylpiperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 7, ZINC ION | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
6H86
 
 | |
6H38
 
 | | The crystal structure of human carbonic anhydrase VII in complex with 4-[(4-fluorophenyl)methyl]-1-piperazinyl]benzenesulfonamide. | | Descriptor: | 4-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 7, GLYCEROL, ... | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
6QXZ
 
 | | Solution structure of the ASHH2 CW domain with the N-terminal histone H3 tail mimicking peptide monomethylated on lysine 4 | | Descriptor: | ALA-ARG-THR-MLZ-GLN-THR-ALA-ARG-TYR, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Dobrovolska, O, Madeleine, N, Teigen, K, Halskau, O, Bril'kov, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Arabidopsis (ASHH2) CW domain binds monomethylated K4 of the histone H3 tail through conformational selection.
Febs J., 287, 2020
|
|
6H34
 
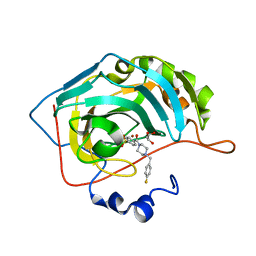 | | The crystal structure of human carbonic anhydrase II in complex with 4-[(4-fluorophenyl)methyl]-1-piperazinyl]benzenesulfonamide. | | Descriptor: | 4-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
6GWV
 
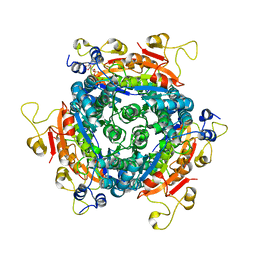 | | Molybdenum storage protein without polymolybdate clusters and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Molybdenum storage protein subunit alpha, ... | | Authors: | Ermler, U, Poppe, J, Bruenle, S. | | Deposit date: | 2018-06-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
6GX4
 
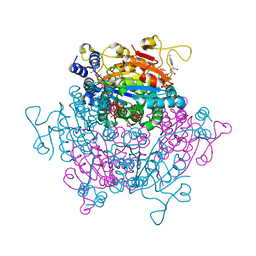 | | The molybdenum storage protein: with ATP/Mn2+ and with POM clusters formed under in vitro conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, MOLYBDATE ION, ... | | Authors: | Poppe, J, Bruenle, S, Hail, R, Ermler, E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
5N0H
 
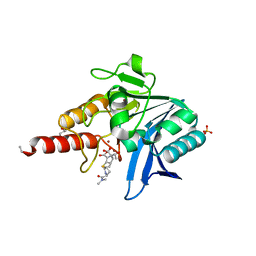 | | Crystal structure of NDM-1 in complex with hydrolyzed meropenem - new refinement | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
3F5E
 
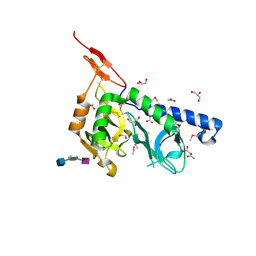 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 2'F-3'SiaLacNAc1-3 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
3F5A
 
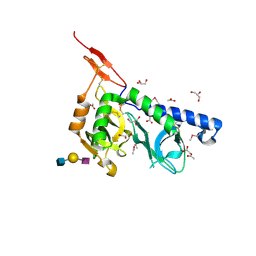 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 3'SiaLacNAc1-3 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
9C2Y
 
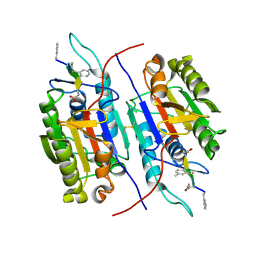 | |
3F53
 
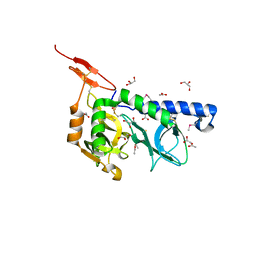 | |
6HQI
 
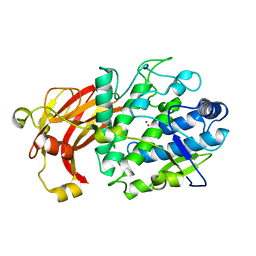 | | holo-form of polyphenol oxidase from Solanum lycopersicum | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Polyphenol oxidase A, ... | | Authors: | Kampatsikas, I, Bijelic, A, Rompel, A. | | Deposit date: | 2018-09-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of tomato polyphenol oxidases provide novel insights into their substrate specificity.
Sci Rep, 9, 2019
|
|
6NOV
 
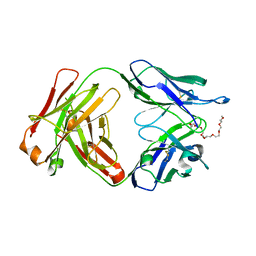 | | A Fab derived from ixekizumab | | Descriptor: | DODECAETHYLENE GLYCOL, Fab Heavy Chain, Fab Light Chain | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Schmitt, M, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
6G4C
 
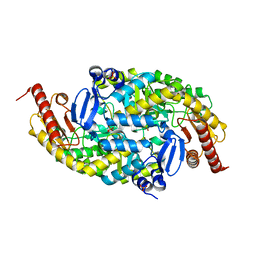 | |
6P90
 
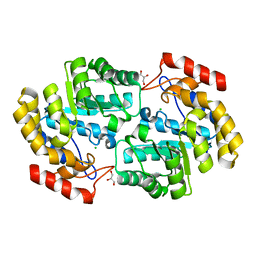 | | Crystal structure of PaDHDPS2-H56Q mutant | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, GLYCEROL | | Authors: | Impey, R.E, Panjikar, S, Hall, C.J, Bock, L.J, Sutton, J.M, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation.
Febs J., 287, 2020
|
|
6G2X
 
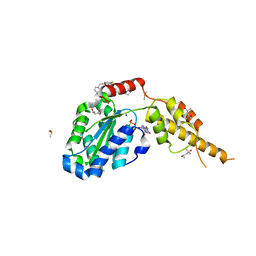 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6GDW
 
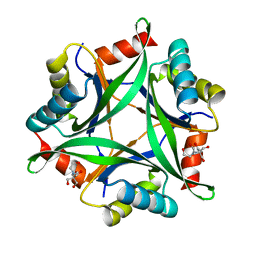 | |
6GDV
 
 | | Structure of CutA from Synechococcus elongatus PCC7942 complexed with Bis-Tris molecule | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
5N0I
 
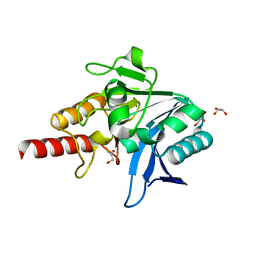 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5NBK
 
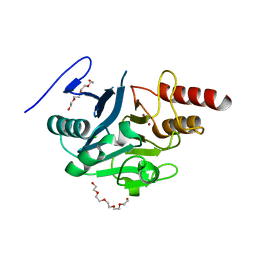 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
6FTD
 
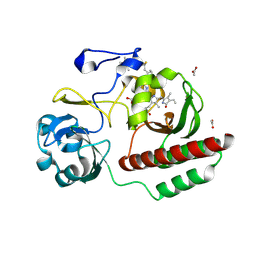 | | Deinococcus radiodurans BphP PAS-GAF H290T mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, ... | | Authors: | Edlund, P, Takala, H, Westenhoff, S, Ihalainen, J.A. | | Deposit date: | 2018-02-21 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Coordination of the biliverdin D-ring in bacteriophytochromes.
Phys Chem Chem Phys, 20, 2018
|
|
6S0V
 
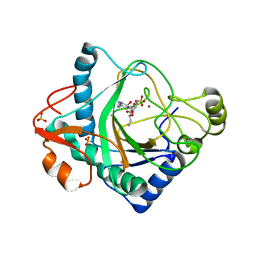 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, neamine and sulfate | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S0S
 
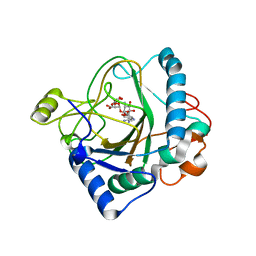 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, ribostamycin B and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, Kanamycin B dioxygenase, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
