7C9P
 
 | | Crystal structure of rice histone-fold dimer GHD8/OsNF-YC2 | | Descriptor: | Nuclear transcription factor Y subunit B-11, Nuclear transcription factor Y subunit C-2 | | Authors: | Shen, C, Liu, H, Guan, Z, Xing, Y, Yin, P. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into DNA Recognition by CCT/NF-YB/YC Complexes in Plant Photoperiodic Flowering.
Plant Cell, 32, 2020
|
|
1M16
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag and Leu 44 Replaced with Phe (L44F), Leu 73 Replaced with Val (L73V), Val 109 Replaced with Leu (V109L) and Cys 117 Replaced with Val (C117V). | | Descriptor: | FORMIC ACID, SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Kim, J, Spielmann, G.L, Logan, T.M, Blaber, M. | | Deposit date: | 2002-06-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold
Protein Sci., 12, 2003
|
|
2DY5
 
 | | Crystal structure of rat heme oxygenase-1 in complex with heme and 2-[2-(4-chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-1,3-dioxolane | | Descriptor: | 1-({2-[2-(4-CHLOROPHENYL)ETHYL]-1,3-DIOXOLAN-2-YL}METHYL)-1H-IMIDAZOLE, CHLORIDE ION, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Takahashi, H, Fukuyama, K. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystallographic and biochemical characterization of the inhibitory action of an imidazole-dioxolane compound on heme oxygenase
Biochemistry, 46, 2007
|
|
8OID
 
 | | Cryo-EM structure of ADP-bound, filamentous beta-actin harboring the N111S mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1CQQ
 
 | | TYPE 2 RHINOVIRUS 3C PROTEASE WITH AG7088 INHIBITOR | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, TYPE 2 RHINOVIRUS 3C PROTEASE | | Authors: | Matthews, D, Ferre, R.A. | | Deposit date: | 1999-08-10 | | Release date: | 1999-09-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8OI8
 
 | | Cryo-EM structure of ADP-bound, filamentous beta-actin harboring the R183W mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8K6Z
 
 | | NMR structure of human leptin | | Descriptor: | Leptin | | Authors: | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
1CU1
 
 | | CRYSTAL STRUCTURE OF AN ENZYME COMPLEX FROM HEPATITIS C VIRUS | | Descriptor: | PHOSPHATE ION, PROTEIN (PROTEASE/HELICASE NS3), ZINC ION | | Authors: | Yao, N, Weber, P.C. | | Deposit date: | 1999-08-20 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase.
Structure Fold.Des., 7, 1999
|
|
6UIB
 
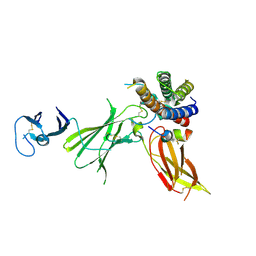 | | Crystal structure of IL23 bound to peptide 23-652 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Durbin, J.D, Wang, J, Afshar, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Integration of phage and yeast display platforms: A reliable and cost effective approach for binning of peptides as displayed on-phage.
Plos One, 15, 2020
|
|
2XPP
 
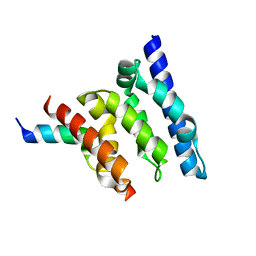 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form III | | Descriptor: | CHROMATIN STRUCTURE MODULATOR, IWS1 | | Authors: | Diebold, M.-L, Koch, M, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
5T1Z
 
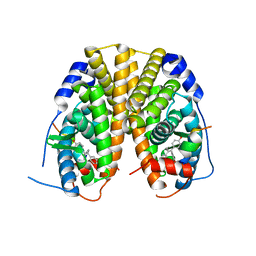 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Ethoxytriphenylethylene and GRIP Peptide | | Descriptor: | 4,4'-[(1Z)-1-(4-ethoxyphenyl)but-1-ene-1,2-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Fanning, S.W, Rajan, S.S, Maximov, P.Y, Abderrahman, B.H, Surojeet, S, Fernandes, D.J, Fan, P, Curpan, R.F, Greene, G.L, Jordan, V.C. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Endoxifen, 4-Hydroxytamoxifen and an Estrogenic Derivative Modulate Estrogen Receptor Complex Mediated Apoptosis in Breast Cancer.
Mol. Pharmacol., 94, 2018
|
|
7BJL
 
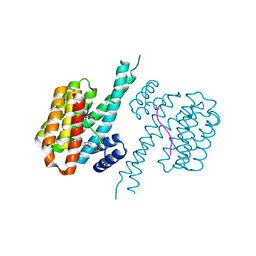 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-142 | | Descriptor: | 14-3-3 protein sigma, 3-[(3~{S})-3-methoxypiperidin-1-yl]-4-nitro-benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJW
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-154 | | Descriptor: | 14-3-3 protein sigma, 4-nitro-3-(4-oxidanylpiperidin-1-yl)benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BGQ
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1019 | | Descriptor: | 14-3-3 protein sigma, 2-methoxy-4-(2-phenylimidazol-1-yl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BJF
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-069 | | Descriptor: | (5-methyl-2-nitro-phenyl) cyclobutanecarboxylate, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BIQ
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-180 | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{R})-2-methyl-3,4-dihydro-2~{H}-quinolin-1-yl]sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BKH
 
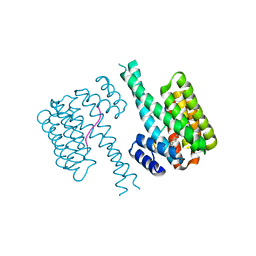 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-153 | | Descriptor: | 14-3-3 protein sigma, 4-nitro-3-[(3S)-3-oxidanylpiperidin-1-yl]benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BGR
 
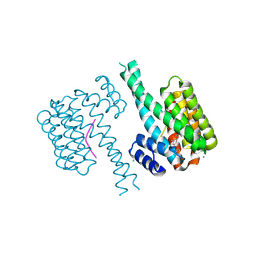 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1016 | | Descriptor: | 14-3-3 protein sigma, 2-methyl-4-(2-phenylimidazol-1-yl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BDP
 
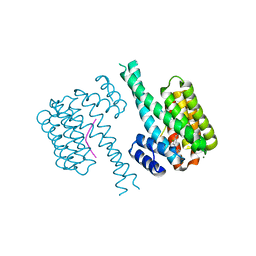 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1017 | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-4-(2-phenylimidazol-1-yl)benzaldehyde, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BDT
 
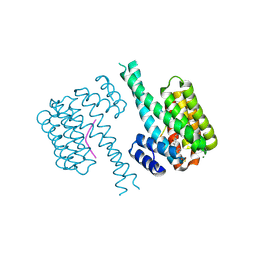 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1009 | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-(2-phenylimidazol-1-yl)benzaldehyde, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BGV
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1012 | | Descriptor: | 14-3-3 protein sigma, 3-methoxy-4-(2-phenylimidazol-1-yl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BDY
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2068B | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-[2-(5-bromanyl-2-fluoranyl-phenyl)imidazol-1-yl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BFW
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2068A | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BI3
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-179 | | Descriptor: | 14-3-3 protein sigma, 4-[(6-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BG3
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2046 | | Descriptor: | 1-(3-bromanyl-4-methyl-phenyl)-2-(2-bromophenyl)imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
