1GT3
 
 | | Complex of Bovine Odorant Binding Protein with dihydromyrcenol | | Descriptor: | (3S)-1-octen-3-ol, 2,6-DIMETHYL-7-OCTEN-2-OL, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1GXA
 
 | |
1GT1
 
 | | Complex of Bovine Odorant Binding Protein with Aminoanthracene and pyrazine | | Descriptor: | (3S)-1-octen-3-ol, 2-ISOBUTYL-3-METHOXYPYRAZINE, ANTHRACEN-1-YLAMINE, ... | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1G74
 
 | |
1GX8
 
 | |
1FTP
 
 | |
1GM6
 
 | | 3-D STRUCTURE OF A SALIVARY LIPOCALIN FROM BOAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, GLYCEROL, ... | | Authors: | Spinelli, S, Vincent, F, Pelosi, P, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-09-11 | | Release date: | 2002-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Boar Salivary Lipocalin. Three-Dimensional X-Ray Structure and Androsterol/Androstenone Docking Simulations.
Eur.J.Biochem., 269, 2002
|
|
1HBQ
 
 | |
1GT5
 
 | | Complexe of Bovine Odorant Binding Protein with benzophenone | | Descriptor: | DIPHENYLMETHANONE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1EIO
 
 | | ILEAL LIPID BINDING PROTEIN IN COMPLEX WITH GLYCOCHOLATE | | Descriptor: | GLYCOCHOLIC ACID, ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Hamilton, J.A, Sacchettini, J.C, Rueterjans, H. | | Deposit date: | 2000-02-27 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ileal lipid binding protein in complex with glycocholate.
Eur.J.Biochem., 267, 2000
|
|
1EPA
 
 | |
1FEM
 
 | | CRYSTALLOGRAPHIC STUDIES ON COMPLEXES BETWEEN RETINOIDS AND PLASMA RETINOL-BINDING PROTEIN | | Descriptor: | RETINOIC ACID, RETINOL BINDING PROTEIN | | Authors: | Zanotti, G, Marcello, M, Malpeli, G, Sartori, G, Berni, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on complexes between retinoids and plasma retinol-binding protein.
J.Biol.Chem., 269, 1994
|
|
1HMS
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1HMR
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | 9-OCTADECENOIC ACID, MUSCLE FATTY ACID BINDING PROTEIN | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1I04
 
 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN-I FROM MOUSE LIVER | | Descriptor: | MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of pheromone binding to mouse major urinary protein (MUP-I)
Protein Sci., 10, 2001
|
|
1I06
 
 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN (MUP-I) COMPLEXED WITH SEC-BUTYL-THIAZOLINE | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Pheromone Binding to Mouse Major Urinary Protein (MUP-I)
Protein Sci., 10, 2001
|
|
1I05
 
 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN (MUP-I) COMPLEXED WITH HYDROXY-METHYL-HEPTANONE | | Descriptor: | 6-HYDROXY-6-METHYL-HEPTAN-3-ONE, CADMIUM ION, MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Pheromone Binding to Mouse Major Urinary Protein (MUP-I)
Protein Sci., 10, 2001
|
|
1IW2
 
 | | X-ray structure of Human Complement Protein C8gamma at pH=7.O | | Descriptor: | Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
1IIU
 
 | |
1IFC
 
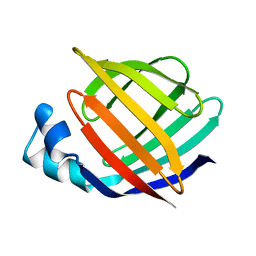 | |
1JBH
 
 | | Solution structure of cellular retinol binding protein type-I in the ligand-free state | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-06-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of Apo- and holo-cellular retinol-binding protein in solution.
J.Biol.Chem., 277, 2002
|
|
1JJJ
 
 | | SOLUTION STRUCTURE OF RECOMBINANT HUMAN EPIDERMAL-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | EPIDERMAL-TYPE FATTY ACID BINDING PROTEIN (E-FABP) | | Authors: | Gutierrez-Gonzalez, L.H, Ludwig, C, Hohoff, C, Rademacher, M, Hanhoff, T, Rueterjans, H, Spener, F, Luecke, C. | | Deposit date: | 2001-07-06 | | Release date: | 2002-06-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human epidermal-type fatty
acid-binding protein (E-FABP)
BIOCHEM.J., 364, 2002
|
|
1HQP
 
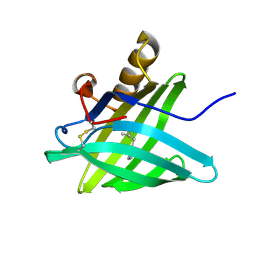 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF PORCINE ODORANT-BINDING PROTEIN | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, ODORANT-BINDING PROTEIN | | Authors: | Perduca, M, Mancia, F, Del Giorgio, R, Monaco, H.L. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a truncated form of porcine odorant-binding protein.
Proteins, 42, 2001
|
|
1HMT
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, STEARIC ACID | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1HN2
 
 | | CRYSTAL STRUCTURE OF BOVINE OBP COMPLEXED WITH AMINOANTHRACENE | | Descriptor: | (3R)-oct-1-en-3-ol, ANTHRACEN-1-YLAMINE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
