5EYG
 
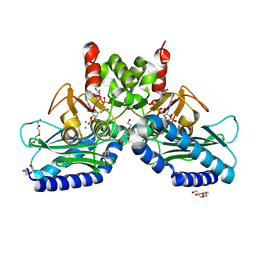 | | Crystal structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
9GQR
 
 | | Structure of a consensus-designed decarboxylase (PSC1) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, decarboxylase | | Authors: | Gavira, J.A, Ramos, J.L, Garcia-Franco, A, de la Torre, J. | | Deposit date: | 2024-09-09 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sustainable synthesis of styrene exploiting a consensus-designed decarboxylase. Purification and crystal structure of PSC1
Mbio, 2025
|
|
7RVV
 
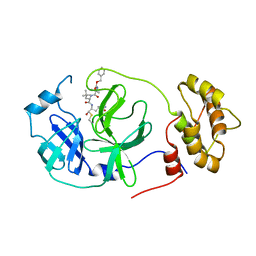 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI22 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-2-methyl-L-alanyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
1M35
 
 | | Aminopeptidase P from Escherichia coli | | Descriptor: | AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Graham, S.C, Lee, M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-06-27 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthorhombic form of Escherichia coli aminopeptidase P at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
9GA5
 
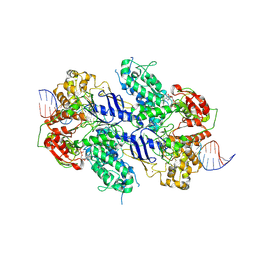 | | MtUvrA2 bound to endogenous E. coli DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endogenous E. coli DNA, UvrABC system protein A, ... | | Authors: | Genta, M, Capelli, R, Ferrara, G, Rizzi, M, Rossi, F, Jeruzalmi, D, Bolognesi, M, Chaves-Sanjuan, A, Miggiano, R. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic understanding of UvrA damage detection and lesion hand-off to UvrB in Nucleotide Excision Repair.
Nat Commun, 16, 2025
|
|
1M41
 
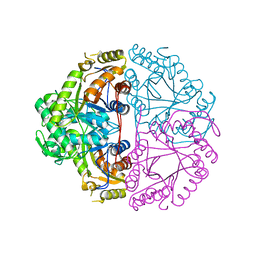 | | Crystal structure of Escherichia coli alkanesulfonate monooxygenase SsuD at 2.3 A resolution | | Descriptor: | FMNH2-dependent alkanesulfonate monooxygenase | | Authors: | Eichhorn, E, Davey, C.A, Sargent, D.F, Leisinger, T, Richmond, T.J. | | Deposit date: | 2002-07-02 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli Alkanesulfonate Monooxygenase SsuD
J.mol.biol., 324, 2002
|
|
1M56
 
 | | Structure of cytochrome c oxidase from Rhodobactor sphaeroides (Wild Type) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Svensson-Ek, M, Abramson, J, Larsson, G, Tornroth, S, Brezezinski, P, Iwata, S. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides.
J.Mol.Biol., 321, 2002
|
|
7RVZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI26 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
9G1W
 
 | |
9GRZ
 
 | | Cryo-EM structure of human SLC35B1 with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Solute carrier family 35 member B1 | | Authors: | Gulati, A, Ahn, D, Suades, A, Drew, D. | | Deposit date: | 2024-09-13 | | Release date: | 2025-05-21 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise ATP translocation into the endoplasmic reticulum by human SLC35B1.
Nature, 643, 2025
|
|
1M4D
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside 2'-N-acetyltransferase, COENZYME A, ... | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
7RVU
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI21 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-isovalyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
9G65
 
 | |
7RVQ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI16 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
1M6R
 
 | |
7RVX
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI24 | | Descriptor: | 3C-like proteinase, benzyl [(1S)-1-cyclopropyl-2-{[(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]amino}-2-oxoethyl]carbamate | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
9GRR
 
 | |
1M6Y
 
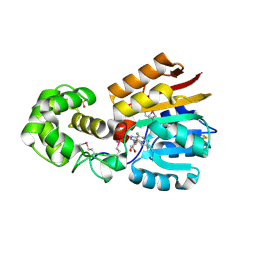 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | Authors: | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-07-17 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
9FYV
 
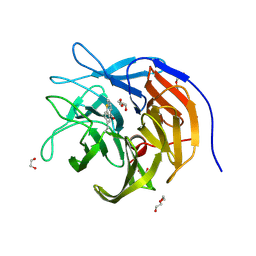 | |
1M7G
 
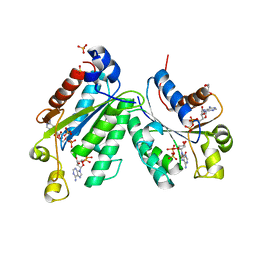 | | Crystal structure of APS kinase from Penicillium Chrysogenum: Ternary structure with ADP and APS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE-2',3'-VANADATE, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Lansdon, E.B, Segel, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
7RW1
 
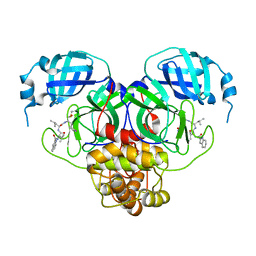 | |
7RML
 
 | | Neisseria meningitidis Methylenetetrahydrofolate reductase in complex with FAD | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pederick, J.L, Wegener, K.L, Salaemae, W, Bruning, J.B. | | Deposit date: | 2021-07-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of meningococcal methylenetetrahydrofolate reductase.
Protein Sci., 32, 2023
|
|
9G0G
 
 | | BsCdaA in complex with Compound 7 | | Descriptor: | CHLORIDE ION, Cyclic di-AMP synthase CdaA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2024-07-08 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
1M54
 
 | | CYSTATHIONINE-BETA SYNTHASE: REDUCED VICINAL THIOLS | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Taoka, S, Lepore, B.W, Kabil, O, Ojha, S, Ringe, D, Banerjee, R. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HUMAN CYSTATHIONINE BETA-SYNTHASE IS A HEME SENSOR PROTEIN. EVIDENCE THAT THE
REDOX SENSOR IS HEME AND NOT THE VICINAL CYSTEINES IN THE CXXC MOTIF SEEN IN THE CRYSTAL STRUCTURE OF THE TRUNCATED ENZYME
BIOCHEMISTRY, 41, 2002
|
|
9GGP
 
 | | Alpha-1-antitrypsin in complex with the Fab fragment of an anti-polymer antibody | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1-antitrypsin, Fab fragment heavy chain of 2C1 monoclonal antibody, ... | | Authors: | Lowen, S.M, Laffranchi, M, Lomas, D.A, Irving, J.A. | | Deposit date: | 2024-08-13 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | High-resolution characterization of ex vivo AAT polymers by solution-state NMR spectroscopy.
Sci Adv, 11, 2025
|
|
