4C3Q
 
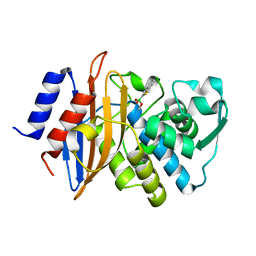 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) at 100K | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, BETA-LACTAMASE TOHO-1 | | Authors: | Coates, L, Tomanicek, S.J, Schrader, T, Weiss, K.L, Ng, J.D, Ostermann, A. | | Deposit date: | 2013-08-26 | | Release date: | 2014-07-09 | | Last modified: | 2017-03-22 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Cryogenic Neutron Protein Crystallography: Routine Methods and Potential Benefits
J.Appl.Crystallogr., 47, 2014
|
|
4BLM
 
 | |
4BD1
 
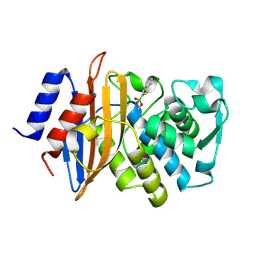 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-16 | | Last modified: | 2017-03-22 | | Method: | NEUTRON DIFFRACTION (2.002 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
4BD0
 
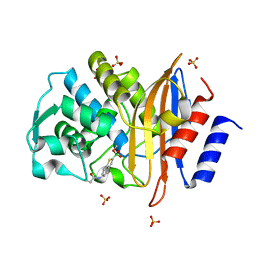 | | X-ray structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, BETA-LACTAMASE TOHO-1, SULFATE ION | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
4A5R
 
 | | Crystal structure of class A beta-lactamase from Bacillus licheniformis BS3 with tazobactam | | Descriptor: | BETA-LACTAMASE, CARBON DIOXIDE, CITRIC ACID, ... | | Authors: | Power, P, Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Class a Beta-Lactamase from Bacillus Licheniformis Inhibited by Tazobactam
To be Published
|
|
3ZYT
 
 | |
3ZNY
 
 | | Crystal structure of the class A extended-spectrum beta-lactamase CTX- M-96, a natural D240G mutant derived from CTX-M-12 | | Descriptor: | CTX-M-12A ENZYME | | Authors: | Power, P, Herman, R, Bouillenne, F, Ghiglione, B, Rodriguez, M.M, Galleni, M, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2013-02-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Kinetic Insights Into the "Ceftazidimase" Behavior of the Extended-Spectrum Beta-Lactamase Ctx-M-96.
Biochemistry, 54, 2015
|
|
3ZHH
 
 | | X-ray structure of the full-length beta-lactamase from M.tuberculosis | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Feiler, C, Fisher, A.C, Marrichi, M.J, Wright, L, Schmidpeter, P.A.M, Blankenfeldt, W, Pavelka, M, DeLisa, M.P. | | Deposit date: | 2012-12-21 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Directed Evolution of Mycobacterium Tuberculosis Beta-Lactamase Reveals Gatekeeper Residue that Regulates Antibiotic Resistance and Catalytic Efficiency.
Plos One, 8, 2013
|
|
3WWX
 
 | | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 | | Descriptor: | OCTANE 1,8-DIAMINE, S12 family peptidase | | Authors: | Arima, J, Nagano, S, Hino, T, Shimone, K, Isoda, Y, Mori, N. | | Deposit date: | 2014-07-03 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 - insight into the structural factors for substrate specificity.
Febs J., 283, 2016
|
|
3WS5
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS4
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2A) | | Descriptor: | Beta-lactamase, CHLORIDE ION, STRONTIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS2
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1C) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS1
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS0
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1A) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRZ
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (without soaking) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRT
 
 | | Wild type beta-lactamase DERIVED FROM CHROMOHALOBACTER SP.560 | | Descriptor: | Beta-lactamase | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W8K
 
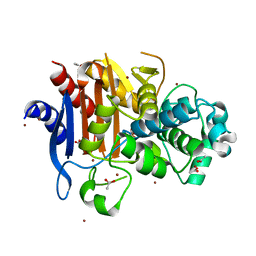 | | Crystal structure of class C beta-lactamase Mox-1 | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Shimizu-ibuka, A, Oguri, T, Furuyama, T, Ishii, Y. | | Deposit date: | 2013-03-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Mox-1, a unique plasmid-mediated class C beta-lactamase with hydrolytic activity towards moxalactam
Antimicrob.Agents Chemother., 58, 2014
|
|
3W4Q
 
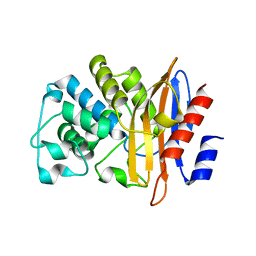 | | Crystal structure of PenA beta-lactamase from Burkholderia multivorans at pH4.2 | | Descriptor: | Beta-lactamase | | Authors: | Nukaga, M, Ohuchi, N, Papp-Wallace, K.M, Taracila, M.A, Bonomo, R.A. | | Deposit date: | 2013-01-10 | | Release date: | 2013-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants
J.Biol.Chem., 288, 2013
|
|
3W4P
 
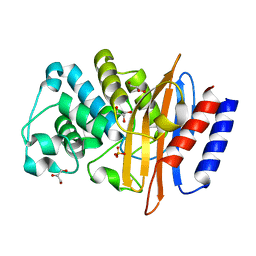 | | Crystal structure of PenI beta-lactamase from Burkholderia pseudomallei at pH7.5 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Nukaga, M, Ohuchi, N, Papp-Wallace, K.M, Taracila, M.A, Bonomo, R.A. | | Deposit date: | 2013-01-10 | | Release date: | 2013-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants
J.Biol.Chem., 288, 2013
|
|
3W4O
 
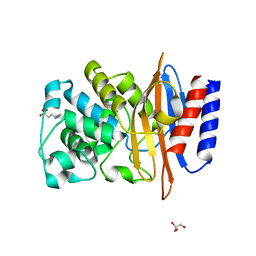 | | Crystal structure of PenI beta-lactamase from Burkholderia pseudomallei at pH9.5 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-lactamase, GLYCEROL | | Authors: | Nukaga, M, Ohuchi, N, Papp-Wallace, K.M, Taracila, M.A, Bonomo, R.A. | | Deposit date: | 2013-01-10 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants
J.Biol.Chem., 288, 2013
|
|
3VWR
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187G/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWQ
 
 | | 6-aminohexanoate-dimer hydrolase S112A/G181D/R187A/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWP
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWN
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187G/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWM
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187A/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
