1B8P
 
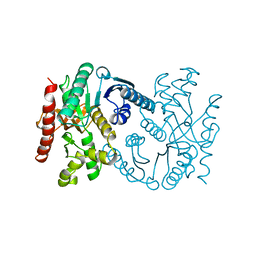 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
4WZC
 
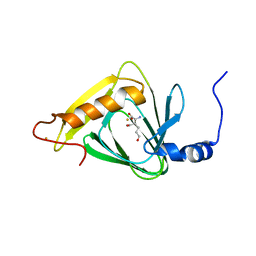 | |
1BA7
 
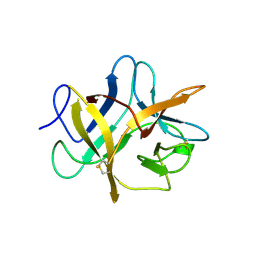 | | SOYBEAN TRYPSIN INHIBITOR | | Descriptor: | TRYPSIN INHIBITOR (KUNITZ) | | Authors: | De Meester, P, Brick, P, Lloyd, L.F, Blow, D.M, Onesti, S. | | Deposit date: | 1998-04-22 | | Release date: | 1998-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Kunitz-type soybean trypsin inhibitor (STI): implication for the interactions between members of the STI family and tissue-plasminogen activator.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
1BEM
 
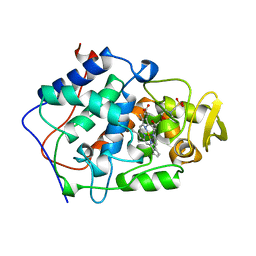 | |
1BEQ
 
 | |
3LTW
 
 | | The structure of mycobacterium marinum arylamine n-acetyltransferase in complex with hydralazine | | Descriptor: | 1-hydrazinophthalazine, Arylamine N-acetyltransferase Nat, FORMIC ACID | | Authors: | Abuhammad, A.M, Lowe, E.D, Fullam, E, Noble, M, Garman, E.F, Sim, E. | | Deposit date: | 2010-02-16 | | Release date: | 2010-07-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the architecture of the Mycobacterium marinum arylamine N-acetyltransferase active site
Protein Cell, 1, 2010
|
|
1BHB
 
 | | Three-dimensional structure of (1-71) bacterioopsin solubilized in methanol-chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Orekhov, V.Y, Pervushin, K.V, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
1BEP
 
 | | EFFECT OF UNNATURAL HEME SUBSTITUTION ON KINETICS OF ELECTRON TRANSFER IN CYTOCHROME C PEROXIDASE | | Descriptor: | YEAST CYTOCHROME C PEROXIDASE, [7-ETHENYL-12-FORMYL-3,8,13,17-TERTRAMETHYL-21H,23H-PORPHINE-2,18-DIPROPANOATO(2)-N21,N22,N23,N24]IRON | | Authors: | Miller, M, Kraut, J. | | Deposit date: | 1998-05-16 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of Unnatural Heme Substitution on Kinetics of Electron Transfer in Cytochrome C Peroxidase
To be Published
|
|
1BEK
 
 | |
4WPK
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form I | | Descriptor: | CITRIC ACID, SODIUM ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
1BES
 
 | |
1BIP
 
 | |
1BEJ
 
 | |
7DGU
 
 | | De novo designed protein H4A1R | | Descriptor: | de novo designed protein H4A1R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
1BIG
 
 | | SCORPION TOXIN BMTX1 FROM BUTHUS MARTENSII KARSCH, NMR, 25 STRUCTURES | | Descriptor: | TOXIN BMTX1 | | Authors: | Blanc, E, Romi-Lebrun, R, Bornet, O, Nakajima, T, Darbon, H. | | Deposit date: | 1998-06-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of two new toxins from the venom of the Chinese scorpion Buthus martensi Karsch blockers of potassium channels.
Biochemistry, 37, 1998
|
|
1BHA
 
 | | THREE-DIMENSIONAL STRUCTURE OF (1-71) BACTERIOOPSIN SOLUBILIZED IN METHANOL-CHLOROFORM AND SDS MICELLES DETERMINED BY 15N-1H HETERONUCLEAR NMR SPECTROSCOPY | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Pervushin, K.V, Orekhov, V.Y, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
3LRI
 
 | | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR I) | | Authors: | Laajoki, L.G, Francis, G.L, Wallace, J.C, Carver, J.A, Keniry, M.A. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I
J.Biol.Chem., 275, 2000
|
|
1B53
 
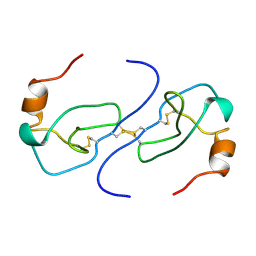 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
1BFG
 
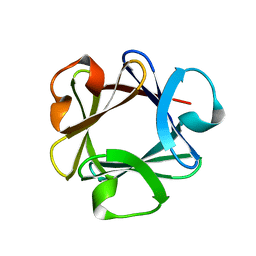 | | CRYSTAL STRUCTURE OF BASIC FIBROBLAST GROWTH FACTOR AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Kitagawa, Y, Ago, H, Katsube, Y, Fujishima, A, Matsuura, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of basic fibroblast growth factor at 1.6 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
7DKK
 
 | | De novo design protein XM2H | | Descriptor: | De novo design protein XM2H | | Authors: | Bin, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
3HY5
 
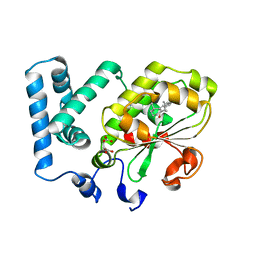 | | Crystal structure of CRALBP | | Descriptor: | L(+)-TARTARIC ACID, RETINAL, Retinaldehyde-binding protein 1 | | Authors: | Stocker, A, He, X, Lobsiger, J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7DKO
 
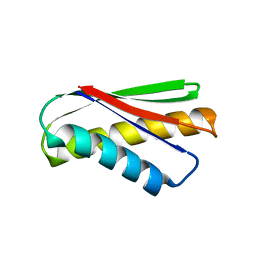 | | De novo design protein AM2M | | Descriptor: | de novo designed protein AM2M | | Authors: | Bin, H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKL
 
 | |
