9B0W
 
 | |
9B11
 
 | |
9B0N
 
 | |
8ZNT
 
 | | Catalytic antibody T99_C220A | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Catalytic antibody T99_C220A, ... | | Authors: | Kobayashi, J, Yoshida, H, Tsuyuguchi, M, Kato, R. | | Deposit date: | 2024-05-28 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and biochemical differences between non-catalytic and catalytic antibodies.
Mabs, 17, 2025
|
|
9AZN
 
 | |
9AZS
 
 | |
9B0O
 
 | |
9AZC
 
 | |
8ZWB
 
 | |
9BE0
 
 | | GC-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*GP*CP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*G)-3'), ZINC ION | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BE1
 
 | | CG-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*GP*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*G)-3'), ZINC ION | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Wang, V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDY
 
 | | AT-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDZ
 
 | | TA-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, ZINC ION, double-stranded DNA | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Wang, V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BKD
 
 | | The structure of human Pdcd4 bound to the 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S12, 40S ribosomal protein S13, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Albacete-Albacete, L, Ramakrishnan, V, S.Fraser, C. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
9BLN
 
 | | The structure of human Pdcd4 bound to the 40S-eIF4A-eIF3-eIF1 complex | | Descriptor: | 40S ribosomal protein S14, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Yifei, D, Albacete-Albacete, L, Ramakrishnan, V, S Fraser, C. | | Deposit date: | 2024-04-30 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
8G3D
 
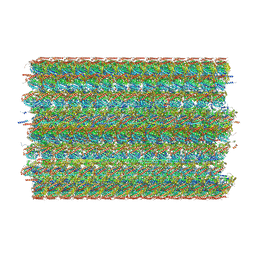 | | 48-nm doublet microtubule from Tetrahymena thermophila strain K40R | | Descriptor: | B2B3_fMIP, B5B6_fMIP, CFAM166A, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins.
Nat Commun, 14, 2023
|
|
8G2Z
 
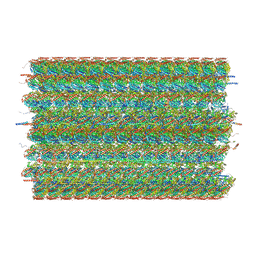 | | 48-nm doublet microtubule from Tetrahymena thermophila strain CU428 | | Descriptor: | B2B3_fMIP, B5B6_fMIP, CFAM166A, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-14 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins.
Nat Commun, 14, 2023
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW7
 
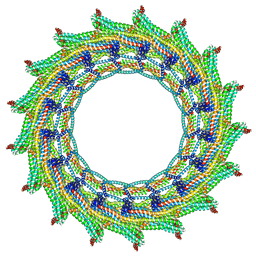 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
7QIJ
 
 | |
7Y4L
 
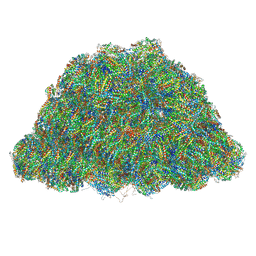 | | PBS of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta 18 subunit, Allophycocyanin beta subunit, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-15 | | Release date: | 2023-01-18 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y5E
 
 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7PE1
 
 | |
7PE2
 
 | |
7ZZ2
 
 | | Cryo-EM structure of "CT pyr" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
