5ZOI
 
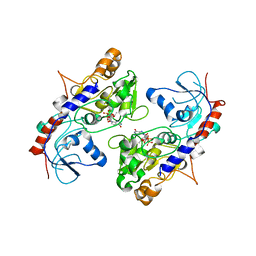 | | Crystal Structure of alpha1,3-Fucosyltransferase | | Descriptor: | Alpha-(1,3)-fucosyltransferase FucT, [[(2S,3R,4S,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5R,6R)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Tan, Y, Yang, G. | | Deposit date: | 2018-04-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Directed evolution of an alpha 1,3-fucosyltransferase using a single-cell ultrahigh-throughput screening method.
Sci Adv, 5, 2019
|
|
4FYQ
 
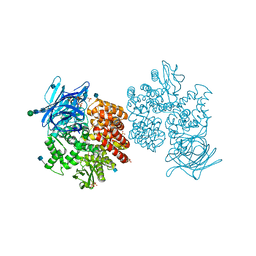 | | Human aminopeptidase N (CD13) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Wong, A.H, Rini, J.M. | | Deposit date: | 2012-07-05 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Crystal Structure of Human Aminopeptidase N Reveals a Novel Dimer and the Basis for Peptide Processing.
J.Biol.Chem., 287, 2012
|
|
3SAQ
 
 | |
5C78
 
 | |
5UQH
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p182 | | Descriptor: | 1,2-ETHANEDIOL, INOSINIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6
To Be Published
|
|
4OEV
 
 | | Crystal structure of NikZ from Campylobacter jejuni in complex with Ni(II) ion | | Descriptor: | GLYCEROL, NICKEL (II) ION, OXALATE ION, ... | | Authors: | Lebrette, H, Cavazza, C. | | Deposit date: | 2014-01-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins.
Structure, 22, 2014
|
|
6QFJ
 
 | |
3JBV
 
 | | Mechanisms of Ribosome Stalling by SecM at Multiple Elongation Steps | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Zhang, J, Pan, X.J, Yan, K.G, Sun, S, Gao, N, Sui, S.F. | | Deposit date: | 2015-10-16 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Mechanisms of ribosome stalling by SecM at multiple elongation steps
Elife, 4, 2015
|
|
4OET
 
 | | Crystal structure of NikZ from Campylobacter jejuni, unliganded form | | Descriptor: | GLYCEROL, Putative peptide ABC-transport system periplasmic peptide-binding protein | | Authors: | Lebrette, H, Cavazza, C. | | Deposit date: | 2014-01-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins.
Structure, 22, 2014
|
|
4OEU
 
 | | Crystal structure of NikZ from Campylobacter jejuni in complex with Ni(L-His) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HISTIDINE, ... | | Authors: | Lebrette, H, Cavazza, C. | | Deposit date: | 2014-01-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins.
Structure, 22, 2014
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
7AXT
 
 | |
5A1J
 
 | | Periplasmic Binding Protein CeuE in complex with ferric 4-LICAM | | Descriptor: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-butane-1,4-diylbis(2,3-dihydroxybenzamide) | | Authors: | Raines, D.J, Moroz, O.V, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2015-04-30 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions of a Periplasmic Binding Protein with a Tetradentate Siderophore Mimic.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6YNC
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with the methylated Fasudil-derived fragment N-methylisoquinoline-5-sulfonamide (soaked) | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ~{N}-methylisoquinoline-5-sulfonamide | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YQI
 
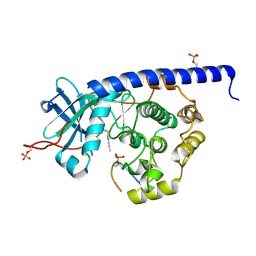 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with long-chain Fasudil-derivative N-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide (soaked) | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ~{N}-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YOT
 
 | |
8S1P
 
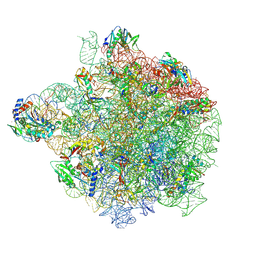 | | YlmH bound to PtRNA-50S | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2024-02-15 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | A role for the S4-domain containing protein YlmH in ribosome-associated quality control in Bacillus subtilis.
Nucleic Acids Res., 52, 2024
|
|
2IPH
 
 | |
7P3K
 
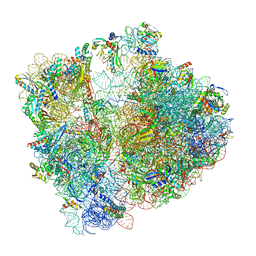 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide (control) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Buschauer, R, Komar, T, Becker, T, Berninghausen, O, Cheng, J, Beckmann, R. | | Deposit date: | 2021-07-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
7RDU
 
 | |
3CEI
 
 | | Crystal Structure of Superoxide Dismutase from Helicobacter pylori | | Descriptor: | FE (III) ION, SULFATE ION, Superoxide dismutase | | Authors: | Esposito, L, Seydel, A, Aiello, R, Sorrentino, G, Cendron, L, Zanotti, G, Zagari, A. | | Deposit date: | 2008-02-29 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the superoxide dismutase from Helicobacter pylori reveals a structured C-terminal extension
Biochim.Biophys.Acta, 1784, 2008
|
|
3AAG
 
 | | Crystal structure of C. jejuni pglb C-terminal domain | | Descriptor: | CALCIUM ION, General glycosylation pathway protein | | Authors: | Maita, N, Kohda, D. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative structural biology of Eubacterial and Archaeal oligosaccharyltransferases.
J.Biol.Chem., 285, 2010
|
|
6OO8
 
 | | Dehaloperoxidase B in complex with substrate pentachlorophenol | | Descriptor: | 1,2-ETHANEDIOL, Dehaloperoxidase B, PENTACHLOROPHENOL, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The multifunctional globin dehaloperoxidase strikes again: Simultaneous peroxidase and peroxygenase mechanisms in the oxidation of EPA pollutants.
Arch.Biochem.Biophys., 673, 2019
|
|
5W17
 
 | |
7TI1
 
 | | Structure of AmpC bound to RPX-7063 at 2.0A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Abendroth, J, Edwards, T.E, Hecker, S.J. | | Deposit date: | 2022-01-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
