5FO8
 
 | | Crystal Structure of Human Complement C3b in Complex with MCP (CCP1-4) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5QB2
 
 | | OXA-48 IN COMPLEX WITH COMPOUND 38 | | Descriptor: | 1,2-ETHANEDIOL, 3-quinolin-6-yl-5-quinolin-7-yl-benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
3IIF
 
 | | Crystal structure of the macro domain of human histone macroH2A1.1 in complex with ADP-ribose (form B) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Core histone macro-H2A.1, Isoform 1 | | Authors: | Hothorn, M, Bortfeld, M, Ladurner, A.G, Scheffzek, K. | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1OFB
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE-REGULATED 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH MANGANESE(II) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Koenig, V, Pfeil, A, Heinrich, G, Braus, G.H, Schneider, T.R. | | Deposit date: | 2003-04-09 | | Release date: | 2004-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Substrate and Metal Complexes of 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase from Saccharomyces Cerevisiae Provide New Insights Into the Catalytic Mechanism
J.Mol.Biol., 337, 2004
|
|
4OGN
 
 | | Co-Crystal Structure of MDM2 with Inhbitor Compound 3 | | Descriptor: | 6-{[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4FST
 
 | | Crystal Structure of the CHK1 | | Descriptor: | 4-[(6,7-dimethoxy-2,4-dihydroindeno[1,2-c]pyrazol-3-yl)ethynyl]-2-methoxyphenol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
1OIK
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with fe, alphaketoglutarate and 2-ethyl-1-hexanesulfuric acid | | Descriptor: | (2R)-2-ETHYL-1-HEXANESULFONIC ACID, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1GZZ
 
 | | Human Insulin-like growth factor; Hamburg data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-06-10 | | Release date: | 2002-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
4ZNR
 
 | | Crystal structure of Dln1 complexed with Man(alpha1-3)Man | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4C6U
 
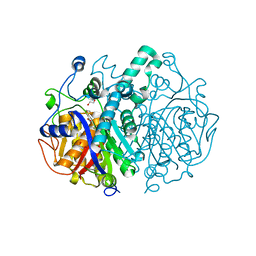 | | Crystal structure of M. tuberculosis KasA in complex with TLM5 | | Descriptor: | (5R)-3-acetyl-4-hydroxy-5-methyl-5-[(1Z)-2-methylbuta-1,3-dien-1-yl]thiophen-2(5H)-one, 1,2-ETHANEDIOL, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, ... | | Authors: | Schiebel, J, Kapilashrami, K, Fekete, A, Bommineni, G.R, Schaefer, C.M, Mueller, M.J, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-09-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Recognition of Mycolic Acid Precursors by Kasa, a Condensing Enzyme and Drug Target from Mycobacterium Tuberculosis
J.Biol.Chem., 288, 2013
|
|
4FZL
 
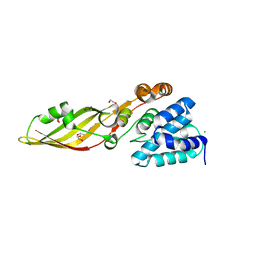 | | High resolution structure of truncated bacteriocin syringacin M from Pseudomonas syringae pv. tomato DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin, CALCIUM ION, ... | | Authors: | Roszak, A.W, Grinter, R, Cogdell, J.R, Walker, D. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The Crystal Structure of the Lipid II-degrading Bacteriocin Syringacin M Suggests Unexpected Evolutionary Relationships between Colicin M-like Bacteriocins.
J.Biol.Chem., 287, 2012
|
|
7EJH
 
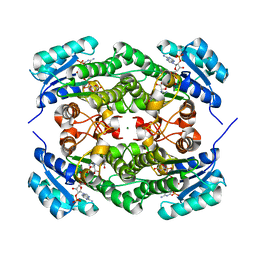 | | Crystal structure of KRED mutant-F147L/L153Q/Y190P/L199A/M205F/M206F and 2-hydroxyisoindoline-1,3-dione complex | | Descriptor: | 2-oxidanylisoindole-1,3-dione, 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72883928 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
3D65
 
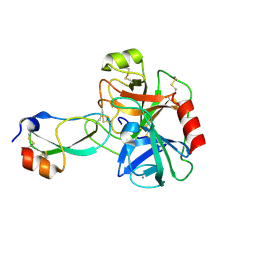 | | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, Textilinin | | Authors: | Millers, E.-K.I, Masci, P.P, Lavin, M.F, de Jersey, J, Guddat, L.W. | | Deposit date: | 2008-05-19 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin
To be Published
|
|
4FSM
 
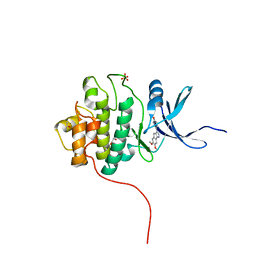 | | Crystal Structure of the CHK1 | | Descriptor: | 4-(6,7-dimethoxy-2,4-dihydroindeno[1,2-c]pyrazol-3-yl)phenol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
2WHL
 
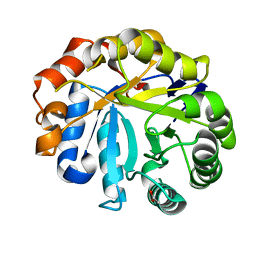 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
3VC1
 
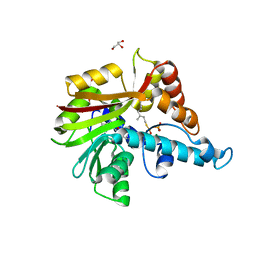 | | Crystal structure of geranyl diphosphate C-methyltransferase from Streptomyces coelicolor A3(2) in complex with Mg2+, geranyl-S-thiolodiphosphate, and S-adenosyl-L-homocysteine | | Descriptor: | GERANYL S-THIOLODIPHOSPHATE, GLYCEROL, Geranyl diphosphate 2-C-methyltransferase, ... | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2012-01-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Geranyl Diphosphate C-Methyltransferase from Streptomyces coelicolor and Implications for the Mechanism of Isoprenoid Modification.
Biochemistry, 51, 2012
|
|
2WVV
 
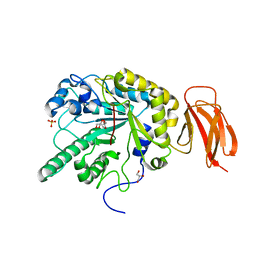 | | Crystal structure of an alpha-L-fucosidase GH29 from Bacteroides thetaiotaomicron | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-L-FUCOSIDASE, GLYCEROL, ... | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach
J.Am.Chem.Soc., 132, 2010
|
|
7HCF
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0000313 | | Descriptor: | (2R)-3-phenyl-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]propan-1-ol, (2S)-3-phenyl-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]propan-1-ol, CHLORIDE ION, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
4NEK
 
 | | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Geffken, K, Chapman, H.C, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1
To be Published
|
|
1OGB
 
 | | Chitinase b from Serratia marcescens mutant D142N | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-29 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
2WQ4
 
 | | N-terminal domain of BC2L-C Lectin from Burkholderia cenocepacia | | Descriptor: | BROMIDE ION, LECTIN, methyl 1-seleno-alpha-L-fucopyranoside | | Authors: | Sulak, O, Cioci, G, Lahman, M, Delia, M, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2009-08-13 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Tnf-Like Trimeric Lectin Domain from Burkholderia Cenocepacia with Specificity for Fucosylated Human Histo-Blood Group Antigens
Structure, 18, 2010
|
|
5QAH
 
 | | OXA-48 IN COMPLEX WITH COMPOUND 9b | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-methoxycarbonylphenyl)benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
3VL1
 
 | | Crystal structure of yeast Rpn14 | | Descriptor: | 26S proteasome regulatory subunit RPN14 | | Authors: | Kim, S, Nishide, A, Saeki, Y, Takagi, K, Tanaka, K, Kato, K, Mizushima, T. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New crystal structure of the proteasome-dedicated chaperone Rpn14 at 1.6 A resolution
Acta Crystallogr.,Sect.F, 68, 2012
|
|
9CT0
 
 | | Native human GABAA receptor of beta2-alpha1-beta2-alpha2-gamma2 assembly | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, J, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2024-07-24 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Resolving native GABA A receptor structures from the human brain.
Nature, 638, 2025
|
|
7HCR
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0000611 | | Descriptor: | (3R)-3-hydroxy-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidine-3-carboxamide, (3S)-3-hydroxy-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
