2RNJ
 
 | | NMR Structure of The S. Aureus VraR DNA Binding Domain | | Descriptor: | Response regulator protein vraR | | Authors: | Donaldson, L.W. | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the Staphylococcus aureus Response Regulator VraR DNA Binding Domain Reveals a Dynamic Relationship between It and Its Associated Receiver Domain
Biochemistry, 47, 2008
|
|
1BWY
 
 | | NMR STUDY OF BOVINE HEART FATTY ACID BINDING PROTEIN | | Descriptor: | PROTEIN (HEART FATTY ACID BINDING PROTEIN) | | Authors: | Lassen, D, Luecke, C, Kveder, M, Mesgarzadeh, A, Schmidt, J.M, Specht, B, Lezius, A, Spener, F, Rueterjans, H. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of bovine heart fatty-acid-binding protein with bound palmitic acid, determined by multidimensional NMR spectroscopy.
Eur.J.Biochem., 230, 1995
|
|
7A64
 
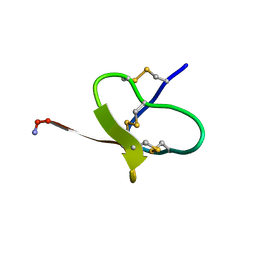 | |
1GH1
 
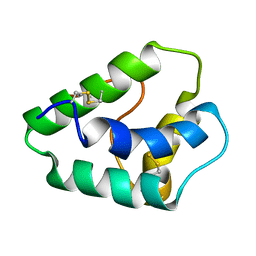 | | NMR STRUCTURES OF WHEAT NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Gincel, E, Simorre, J.P, Caille, A, Marion, D, Ptak, M, Vovelle, F. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers.
Eur.J.Biochem., 226, 1994
|
|
2A7U
 
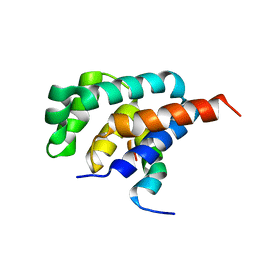 | | NMR solution structure of the E.coli F-ATPase delta subunit N-terminal domain in complex with alpha subunit N-terminal 22 residues | | Descriptor: | ATP synthase alpha chain, ATP synthase delta chain | | Authors: | Wilkens, S, Borchardt, D, Weber, J, Senior, A.E. | | Deposit date: | 2005-07-06 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Interaction of the delta and alpha Subunits of the Escherichia coli F(1)F(0)-ATP Synthase by NMR Spectroscopy
Biochemistry, 44, 2005
|
|
6TPB
 
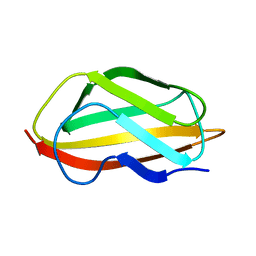 | | NMR structure of the apo-form of Pseudomonas fluorescens CopC | | Descriptor: | Putative copper resistance protein | | Authors: | Persson, K.C, Mayzel, M, Karlsson, B.G, Peciulyte, A, Olsson, L, Wittung Stafshede, P, Salomon Johansen, K, Horvath, I. | | Deposit date: | 2019-12-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Pseudomonas fluorescens CopC
To Be Published
|
|
5T43
 
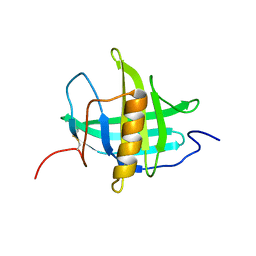 | |
5U87
 
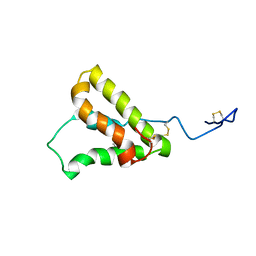 | | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin | | Descriptor: | Preproalbumin PawS1 | | Authors: | Franke, B, James, A.M, Mobli, M, Colgrave, M.L, Mylne, J.S, Rosengren, K.J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin
to be published
|
|
5XQ5
 
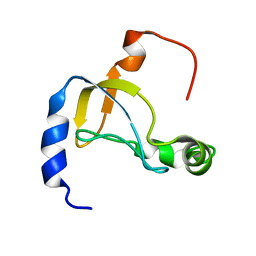 | |
2RV8
 
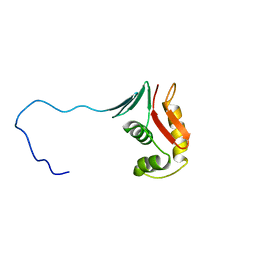 | |
1N4C
 
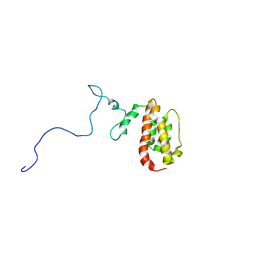 | | NMR Structure of the J-Domain and Clathrin Substrate Binding Domain of Bovine Auxilin | | Descriptor: | Auxilin | | Authors: | Gruschus, J.M, Han, C.J, Greener, T, Greene, L.E, Ferretti, J.A, Eisenberg, E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional fragment of auxilin required for catalytic uncoating of clathrin-coated vesicles.
Biochemistry, 43, 2004
|
|
8QDS
 
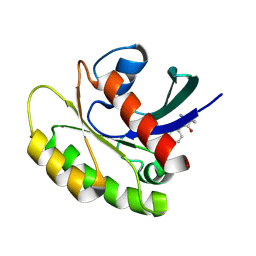 | |
1N91
 
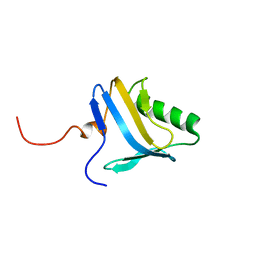 | | Solution NMR Structure of Protein yggU from Escherichia coli. Northeast Structural Genomics Consortium Target ER14. | | Descriptor: | orf, hypothetical protein | | Authors: | Aramini, J.M, Xiao, R, Huang, Y.J, Acton, T.B, Wu, M.J, Mills, J.L, Tejero, R.T, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-11-21 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Resonance assignments for the hypothetical protein yggU from Escherichia coli.
J.Biomol.Nmr, 27, 2003
|
|
3BTB
 
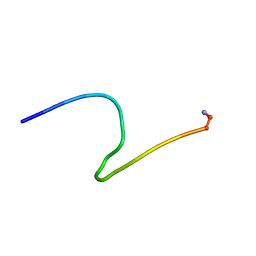 | |
1Q53
 
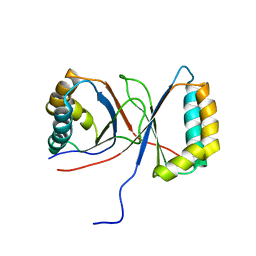 | |
2RN7
 
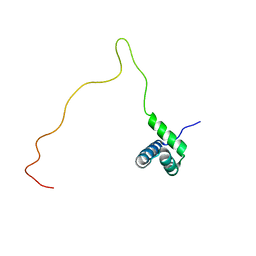 | | NMR solution structure of TnpE protein from Shigella flexneri. Northeast Structural Genomics Target SfR125 | | Descriptor: | IS629 orfA | | Authors: | Ramelot, T.A, Cort, J.R, Semesi, A, Garcia, M, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-08 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of TnpE protein from Shigella flexneri. Northeast
Structural Genomics Target SfR125
To be Published
|
|
1M7L
 
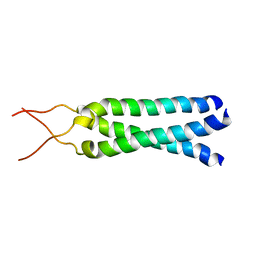 | | Solution Structure of the Coiled-Coil Trimerization Domain from Lung Surfactant Protein D | | Descriptor: | Pulmonary surfactant-associated protein D | | Authors: | Kovacs, H, O'Donoghue, S.I, Hoppe, H.-J, Comfort, D, Reid, K.B.M, Campbell, I.D, Nilges, M. | | Deposit date: | 2002-07-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the coiled-coil trimerization domain from lung surfactant protein D
J.BIOMOL.NMR, 24, 2002
|
|
1PLX
 
 | |
1LJZ
 
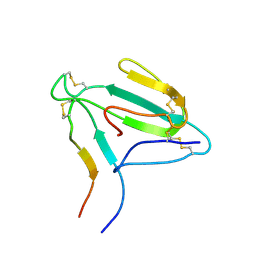 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Eisenstein, M, Chill, J.H, Anglister, J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-07-17 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1L4W
 
 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Rodriguez, E, Eisenstein, M, Anglister, J. | | Deposit date: | 2002-03-06 | | Release date: | 2002-07-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1PRS
 
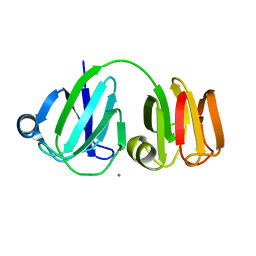 | | NMR-DERIVED THREE-DIMENSIONAL SOLUTION STRUCTURE OF PROTEIN S COMPLEXED WITH CALCIUM | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Bagby, S, Harvey, T.S, Eagle, S.G, Inouye, S, Ikura, M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR-derived three-dimensional solution structure of protein S complexed with calcium.
Structure, 2, 1994
|
|
1K1Z
 
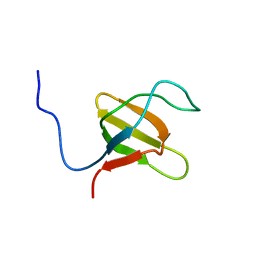 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
1PLW
 
 | |
1L3O
 
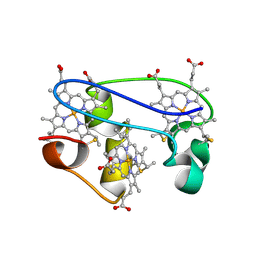 | | SOLUTION STRUCTURE DETERMINATION OF THE FULLY OXIDIZED DOUBLE MUTANT K9-10A CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS, ENSEMBLE OF 35 STRUCTURES | | Descriptor: | HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Turano, P, Bruschi, M, Durand, M.C, Giudici-Orticoni, M.T, Dolla, A. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A quick solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans and mechanistic implications.
J.Biomol.NMR, 22, 2002
|
|
1PRR
 
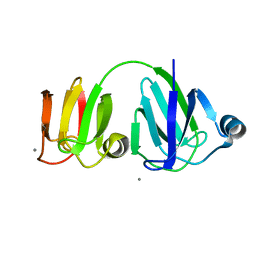 | | NMR-DERIVED THREE-DIMENSIONAL SOLUTION STRUCTURE OF PROTEIN S COMPLEXED WITH CALCIUM | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Bagby, S, Harvey, T.S, Eagle, S.G, Inouye, S, Ikura, M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR-derived three-dimensional solution structure of protein S complexed with calcium.
Structure, 2, 1994
|
|
