4AD9
 
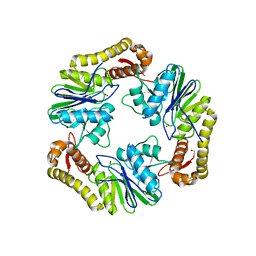 | | Crystal structure of human LACTB2. | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE-LIKE PROTEIN 2, ZINC ION | | Authors: | Allerston, C.K, Krojer, T, Shrestha, B, Burgess Brown, N, Chalk, R, Elkins, J.M, Filippakopoulos, P, Pike, A.C.W, Muniz, J.R.C, Vollmar, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Lactb2, a Metallo-Beta-Lactamase Protein, as a Human Mitochondrial Endoribonuclease
Nucleic Acids Res., 44, 2016
|
|
4AXH
 
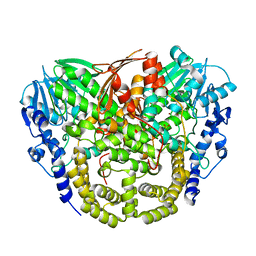 | | Structure and mechanism of the first inverting alkylsulfatase specific for secondary alkylsulfatases | | Descriptor: | SEC-ALKYLSULFATASE, SULFATE ION, ZINC ION | | Authors: | Knaus, T, Schober, M, Faber, K, Macheroux, P, Wagner, U.G. | | Deposit date: | 2012-06-13 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Mechanism of an Inverting Alkylsulfatase from Pseudomonas Sp. Dsm6611 Specific for Secondary Alkylsulfates.
FEBS J., 279, 2012
|
|
2QED
 
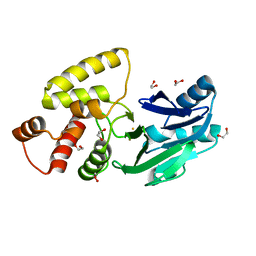 | | Crystal structure of Salmonella thyphimurium LT2 glyoxalase II | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Hydroxyacylglutathione hydrolase | | Authors: | Leite, N.R, Campos Bermudez, V.A, Krogh, R, Oliva, G, Soncini, F.C, Vila, A.J. | | Deposit date: | 2007-06-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and Structural Characterization of Salmonella typhimurium Glyoxalase II: New Insights into Metal Ion Selectivity
Biochemistry, 46, 2007
|
|
4PDX
 
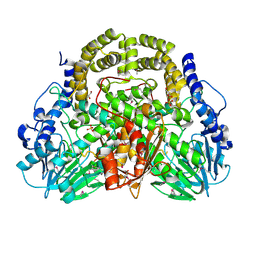 | |
4NUR
 
 | | Crystal structure of thermostable alkylsulfatase SdsAP from Pseudomonas sp. S9 | | Descriptor: | MAGNESIUM ION, PSdsA, ZINC ION | | Authors: | Sun, L.F, Su, Y.T, Zhao, Y.H, Cai, Z.X, Wu, Y. | | Deposit date: | 2013-12-04 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Crystal structure of thermostable alkylsulfatase SdsAP from Pseudomonas sp. S9
To be Published
|
|
3IEM
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog | | Descriptor: | CITRATE ANION, RNA (5'-R(*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU))-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitus, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog
To be Published
|
|
3IEL
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP
To be Published
|
|
3IEK
 
 | | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IE1
 
 | | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA | | Descriptor: | CITRATE ANION, RNA (5'-R(P*UP*UP*UP*U)-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA
To be Published
|
|
3IE2
 
 | | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IDZ
 
 | | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IE0
 
 | | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
2XR1
 
 | | DIMERIC ARCHAEAL CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR WITH N-TERMINAL KH DOMAINS (KH-CPSF) FROM METHANOSARCINA MAZEI | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 100 KD SUBUNIT, ZINC ION | | Authors: | Mir-Montazeri, B, Ammelburg, M, Forouzan, D, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Dimeric Archaeal Cleavage and Polyadenylation Specificity Factor.
J.Struct.Biol., 173, 2011
|
|
2ZO4
 
 | | Crystal structure of metallo-beta-lactamase family protein TTHA1429 from Thermus thermophilus HB8 | | Descriptor: | Metallo-beta-lactamase family protein, ZINC ION | | Authors: | Yamamura, A, Nagata, K, Agari, Y, Ebihara, A, Nakagawa, N, Yokoyama, S, Kuramitsu, S, Tanokura, M. | | Deposit date: | 2008-05-05 | | Release date: | 2009-03-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of TTHA1429, a novel metallo-beta-lactamase superfamily protein from Thermus thermophilus HB8.
Proteins, 73, 2008
|
|
3A4Y
 
 | | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8
to be published
|
|
2YHE
 
 | | Structure determination of the stereoselective inverting sec- alkylsulfatase Pisa1 from Pseudomonas sp. | | Descriptor: | SEC-ALKYL SULFATASE, SULFATE ION, ZINC ION | | Authors: | Kepplinger, B, Faber, K, Macheroux, P, Schober, M, Knaus, T, Wagner, U.G. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Mechanism of an Inverting Alkylsulfatase from Pseudomonas Sp. Dsm6611 Specific for Secondary Alkylsulfates.
FEBS J., 279, 2012
|
|
6K6W
 
 | |
6K6S
 
 | |
6I1D
 
 | | Structure of the Ysh1-Mpe1 nuclease complex from S.cerevisiae | | Descriptor: | Endoribonuclease YSH1, GLYCEROL, Protein MPE1, ... | | Authors: | Hill, C.H, Boreikaite, V, Kumar, A, Casanal, A, Kubik, P, Degliesposti, G, Maslen, S, Mariani, A, von Loeffelholz, O, Girbig, M, Skehel, M, Passmore, L.A. | | Deposit date: | 2018-10-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Activation of the Endonuclease that Defines mRNA 3' Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex.
Mol.Cell, 73, 2019
|
|
7TL5
 
 | | Crystal structure of putative hydrolase yjcS from Klebsiella pneumoniae. | | Descriptor: | 1,2-ETHANEDIOL, Lactamase_B domain-containing protein | | Authors: | Chang, C, Endres, M, Wu, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
2DKF
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8, a RNA Degradation Protein of the Metallo-beta-lactamase Superfamily | | Descriptor: | ZINC ION, metallo-beta-lactamase superfamily protein | | Authors: | Ishikawa, I, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-10 | | Release date: | 2006-12-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of TTHA0252 from Thermus thermophilus HB8, a RNA degradation protein of the metallo-beta-lactamase superfamily
J.Biochem.(Tokyo), 140, 2006
|
|
2CG3
 
 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa. | | Descriptor: | SDSA1, ZINC ION | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Third Class of Sulfatases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CG2
 
 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with sulfate | | Descriptor: | SDSA1, SULFATE ION, ZINC ION | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CFU
 
 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with 1-decane-sulfonic-acid. | | Descriptor: | 1-DECANE-SULFONIC-ACID, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL, ... | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-23 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CFZ
 
 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with 1-dodecanol | | Descriptor: | 1-DODECANOL, DI(HYDROXYETHYL)ETHER, SDS HYDROLASE SDSA1, ... | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-26 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
