2JCB
 
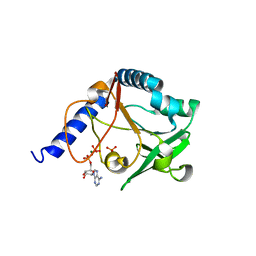 | | The crystal structure of 5-formyl-tetrahydrofolate cycloligase from Bacillus anthracis (BA4489) | | Descriptor: | 5-FORMYLTETRAHYDROFOLATE CYCLO-LIGASE FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meier, C, Carter, L.G, Winter, G, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 5-Formyltetrahydrofolate Cyclo-Ligase from Bacillus Anthracis (Ba4489).
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3II6
 
 | |
1NWI
 
 | |
5COV
 
 | |
5G4Z
 
 | | Structural basis for carboxylic acid recognition by a Cache chemosensory domain. | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with Cache sensor, TRIETHYLENE GLYCOL, UNKNOWN LIGAND | | Authors: | Brewster, J, McKellar, J.L.O, Newman, J, Peat, T.S, Gerth, M.L. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae.
Sci Rep, 6, 2016
|
|
5COT
 
 | |
1P31
 
 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine Ligase (MurC) from Haemophilus influenzae | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--alanine ligase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-16 | | Release date: | 2003-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
5COU
 
 | |
2H89
 
 | | Avian Respiratory Complex II with Malonate Bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
2H88
 
 | | Avian Mitochondrial Respiratory Complex II at 1.8 Angstrom Resolution | | Descriptor: | AZIDE ION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
1P3D
 
 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine ligase (MurC) in Complex with UMA and ANP. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--alanine ligase, ... | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
6D3N
 
 | | Crystal structure of h4-1BB ligand | | Descriptor: | GLYCEROL, Tumor necrosis factor ligand superfamily member 9 | | Authors: | Aruna, B, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
2V7B
 
 | |
1EHI
 
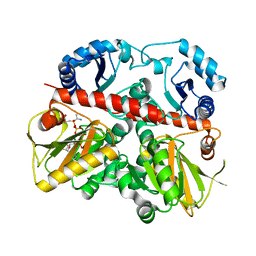 | | D-ALANINE:D-LACTATE LIGASE (LMDDL2) OF VANCOMYCIN-RESISTANT LEUCONOSTOC MESENTEROIDES | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE:D-LACTATE LIGASE, ... | | Authors: | Kuzin, A.P, Sun, T, Jorczak-Baillass, J, Healy, V.L, Walsh, C.T, Knox, J.R. | | Deposit date: | 2000-02-21 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Enzymes of vancomycin resistance: the structure of D-alanine-D-lactate ligase of naturally resistant Leuconostoc mesenteroides.
Structure Fold.Des., 8, 2000
|
|
2DLN
 
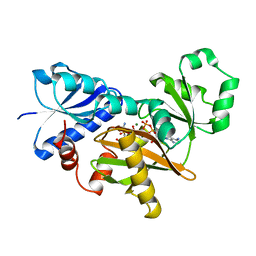 | | VANCOMYCIN RESISTANCE: STRUCTURE OF D-ALANINE:D-ALANINE LIGASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE--D-ALANINE LIGASE, ... | | Authors: | Knox, J.R, Moews, P.C, Fan, C. | | Deposit date: | 1994-07-18 | | Release date: | 1995-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Vancomycin resistance: structure of D-alanine:D-alanine ligase at 2.3 A resolution.
Science, 266, 1994
|
|
6LL9
 
 | |
6M2U
 
 | |
6N0V
 
 | | tRNA ligase | | Descriptor: | MANGANESE (II) ION, tRNA ligase | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
5WFY
 
 | |
6N0T
 
 | | tRNA ligase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
4CC6
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-{[2-(1H-pyrazolo[3,4-c]pyridin-3-yl)-6-(trifluoromethyl)pyridin-4-yl]amino}ethanol, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
3GDE
 
 | | The closed conformation of ATP-dependent DNA ligase from Archaeoglobus fulgidus | | Descriptor: | DNA ligase, PHOSPHATE ION | | Authors: | Kim, D.J, Kim, H.-W, Kim, O, Kim, H.S, Lee, S.J, Suh, S.W. | | Deposit date: | 2009-02-24 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP-dependent DNA ligase from Archaeoglobus fulgidus displays a tightly closed conformation
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3RR5
 
 | | DNA ligase from the archaeon Thermococcus sp. 1519 | | Descriptor: | DNA ligase, MAGNESIUM ION | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Popov, V.O, Polyakov, K.M, Ravin, N.V, Shabalin, I.G, Skryabin, K.G, Stekhanova, T.N, Kovalchuk, M.V. | | Deposit date: | 2011-04-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.018 Å) | | Cite: | ATP-dependent DNA ligase from Thermococcus sp. 1519 displays a new arrangement of the OB-fold domain.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1KKC
 
 | | Crystal structure of Aspergillus fumigatus MnSOD | | Descriptor: | MANGANESE (II) ION, Manganese Superoxide Dismutase | | Authors: | Fluckiger, S, Mittl, P.R.E, Scapozza, L, Fijten, H, Folkers, G, Grutter, M.G, Blaser, K, Crameri, R. | | Deposit date: | 2001-12-07 | | Release date: | 2001-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of the crystal structures of the human manganese superoxide dismutase and the homologous Aspergillus fumigatus allergen at 2-A resolution.
J.Immunol., 168, 2002
|
|
2DEQ
 
 | |
