5CZ6
 
 | | Yeast 20S proteasome beta5-T1A mutant in complex with Syringolin A, propeptide expressed in trans | | Descriptor: | (2S)-2-[[(2S)-1-[[(5S,8S,9E)-2,7-dioxo-5-propan-2-yl-1,6-diazacyclododeca-3,9-dien-8-yl]amino]-3-methyl-1-oxo-butan-2-yl]carbamoylamino]-3-methyl-butanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
2V3I
 
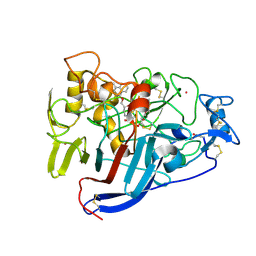 | | Hypocrea jecorina Cel7A in complex with (R)-dihydroxy-phenanthrenolol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(2R)-2-HYDROXY-3-(9-PHENANTHRYLOXY)PROPYL]AMINO}PROPANE-1,3-DIOL, COBALT (II) ION, ... | | Authors: | Fagerstrom, A, Sandgren, M, Berg, U, Stahlberg, J. | | Deposit date: | 2007-06-18 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A Study of the Chiral Recognition Mechanisms of Cellobiohydrolase Cel7A for Ligands Based on the Beta-Blocker Motif: Crystal Structures, Microcalorimetry and Computational Modelling of Cel7A-Inhibitor Complexes.
To be Published
|
|
7GGQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-92e193ae-1 (Mpro-x12692) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
1XMD
 
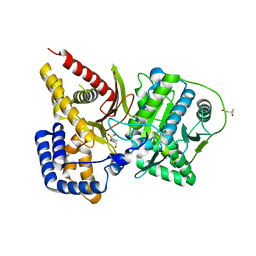 | | M335V mutant structure of mouse carnitine octanoyltransferase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peroxisomal carnitine O-octanoyltransferase | | Authors: | Jogl, G, Hsiao, Y.S, Tong, L. | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity.
J.Biol.Chem., 280, 2005
|
|
4HDK
 
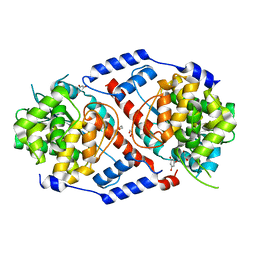 | | Crystal Structure of ArsAB in Complex with Phloroglucinol | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
1GDL
 
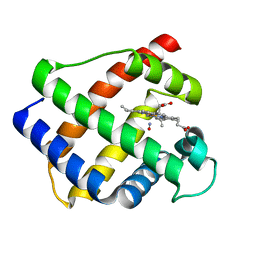 | |
7WLY
 
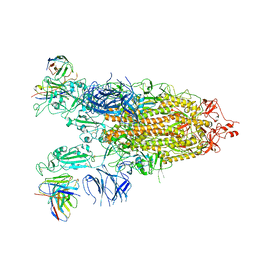 | | Cryo-EM structure of the Omicron S in complex with 35B5 Fab(1 down- and 2 up RBDs) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, ... | | Authors: | Wang, X, Zhu, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | 35B5 antibody potently neutralizes SARS-CoV-2 Omicron by disrupting the N-glycan switch via a conserved spike epitope.
Cell Host Microbe, 30, 2022
|
|
4CBY
 
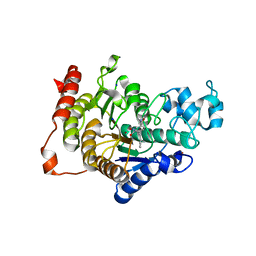 | | Design, synthesis, and biological evaluation of potent and selective Class IIa HDAC inhibitors as a potential therapy for Huntington's disease | | Descriptor: | (1R,2R,3R)-2-[4-(1,3-oxazol-5-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, SODIUM ION, ... | | Authors: | Burli, R.W, Luckhurst, C.A, Aziz, O, Matthews, K.L, Yates, D, Lyons, K.A, Beconi, M, McAllister, G, Breccia, P, Stott, A.J, Penrose, S.D, Wall, M, Lamers, M, Leonard, P, Mueller, I, Richardson, C.M, Jarvis, R, Stones, L, Hughes, S, Wishart, G, Haughan, A.F, O'Connell, C, Mead, T, McNeil, H, Vann, J, Mangette, J, Maillard, M, Beaumont, V, Munoz-Sanjuan, I, Dominguez, C. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease.
J. Med. Chem., 56, 2013
|
|
1TBH
 
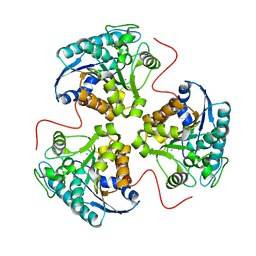 | | H141D mutant of rat liver arginase I | | Descriptor: | Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | Deposit date: | 2004-05-20 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
6ZR4
 
 | |
9MMI
 
 | | Myo-inositol-1(or 4)-monophosphatase that can perform essential dephosphorylation step to facilitate riboflavin biosynthesis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, MAGNESIUM ION, ... | | Authors: | Hoffpauir, Z.A, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2024-12-20 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of the phosphatase essential for riboflavin biosynthesis in Aquifex aeolicus.
J.Biol.Chem., 301, 2025
|
|
7GGN
 
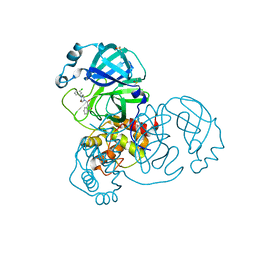 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ERI-UCB-d6de1f3c-1 (Mpro-x12679) | | Descriptor: | 3C-like proteinase, 4-[4-(3-chlorophenyl)-3-oxopiperazine-1-carbonyl]quinolin-2(1H)-one, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2PKH
 
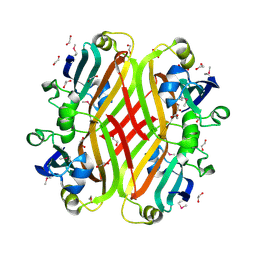 | | Structural Genomics, the crystal structure of the C-terminal domain of histidine utilization repressor from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Histidine utilization repressor | | Authors: | Tan, K, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the C-terminal domain of histidine utilization repressor from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
4YRK
 
 | |
7GGR
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-c7771779-1 (Mpro-x12695) | | Descriptor: | (4S)-6-chloro-4-hydroxy-N-(isoquinolin-4-yl)-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4Y3T
 
 | | Endothiapepsin in complex with fragment 207 | | Descriptor: | (3R)-piperidin-3-yl(piperidin-1-yl)methanone, (3S)-piperidin-3-yl(piperidin-1-yl)methanone, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Huschmann, F.U, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
To Be Published
|
|
7GH1
 
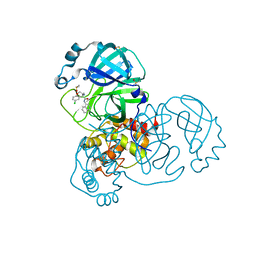 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-2bb0cf2b-1 (Mpro-x12731) | | Descriptor: | (4S)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2Q3G
 
 | | Structure of the PDZ domain of human PDLIM7 bound to a C-terminal extension from human beta-tropomyosin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ and LIM domain protein 7 | | Authors: | Gileadi, C, Papagrigoriou, E, Elkins, J, Burgess-Brown, N, Salah, E, Gileadi, O, Umeano, C, Bunkoczi, G, von Delft, F, Uppenberg, J, Pike, A.C.W, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms
Protein Sci., 19, 2010
|
|
7GCU
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-25 (Mpro-x10787) | | Descriptor: | 1-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)-3-oxocyclobutane-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
1NE6
 
 | | Crystal structure of Sp-cAMP binding R1a subunit of cAMP-dependent protein kinase | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Jones, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2002-12-10 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of RIalpha Subunit of Cyclic Adenosine 5'-Monophosphate (cAMP)-Dependent Protein Kinase Complexed with (R(p))-Adenosine 3',5'-Cyclic Monophosphothioate and (S(p))-Adenosine 3',5'-Cyclic Monophosphothioate, the Phosphothioate Analogues of cAMP.
Biochemistry, 43, 2004
|
|
1FY4
 
 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-ARG, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2FGO
 
 | | Structure of the 2[4FE-4S] ferredoxin from Pseudomonas aeruginosa | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER | | Authors: | Giastas, P, Pinotsis, N, Mavridis, I.M. | | Deposit date: | 2005-12-22 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structure of the 2[4Fe-4S] ferredoxin from Pseudomonas aeruginosa at 1.32-A resolution: comparison with other high-resolution structures of ferredoxins and contributing structural features to reduction potential values.
J.Biol.Inorg.Chem., 11, 2006
|
|
7GDU
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-SGC-a8a902d9-1 (Mpro-x11225) | | Descriptor: | (3R)-3-(4-hydroxypiperidin-1-yl)-N-(4-methylpyridin-3-yl)-3-(thiophen-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5L52
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 14 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
4H03
 
 | | Crystal structure of NAD+-Ia-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-07 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
