8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8HL7
 
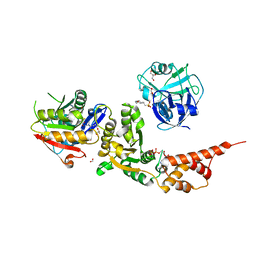 | |
6LXF
 
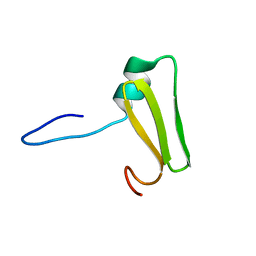 | | Aromatic interactions drive the coupled folding and binding of the intrinsically disordered Sesbania mosaic virus VPg protein. | | Descriptor: | Polyprotein | | Authors: | Dixit, K, Karanth, N.M, Nair, S, Kumari, K, Chakrabarti, K.S, Savithri, H.S, Sarma, S.P. | | Deposit date: | 2020-02-11 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Aromatic Interactions Drive the Coupled Folding and Binding of the Intrinsically Disordered Sesbania mosaic Virus VPg Protein.
Biochemistry, 59, 2020
|
|
5CIC
 
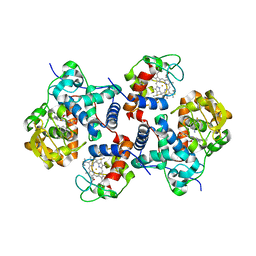 | |
5CIH
 
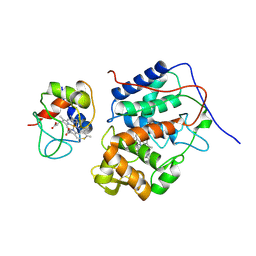 | |
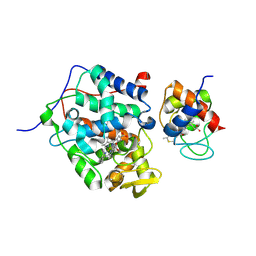
5CIG
 
 | |
5CIB
 
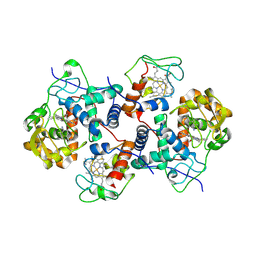 | | Complex of yeast cytochrome c peroxidase (W191G) bound to 2,4-dimethylaniline with iso-1 cytochrome c | | Descriptor: | 2,4-dimethylaniline, Cytochrome c iso-1, Cytochrome c peroxidase, ... | | Authors: | Crane, B.R, Payne, T.M. | | Deposit date: | 2015-07-11 | | Release date: | 2016-08-03 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Constraints on the Radical Cation Center of Cytochrome c Peroxidase for Electron Transfer from Cytochrome c.
Biochemistry, 55, 2016
|
|
2PCP
 
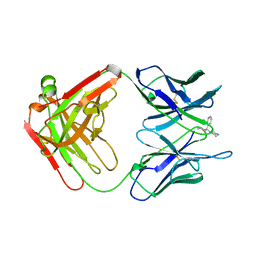 | | ANTIBODY FAB COMPLEXED WITH PHENCYCLIDINE | | Descriptor: | 1-(PHENYL-1-CYCLOHEXYL)PIPERIDINE, IMMUNOGLOBULIN | | Authors: | Lim, K, Owens, S.M, Arnold, L, Sacchettini, J.C, Linthicum, D.S. | | Deposit date: | 1998-06-02 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of monoclonal 6B5 Fab complexed with phencyclidine.
J.Biol.Chem., 273, 1998
|
|
5CIF
 
 | |
5CIE
 
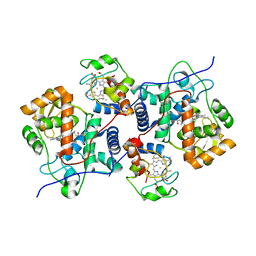 | |
6CBD
 
 | |
2OLB
 
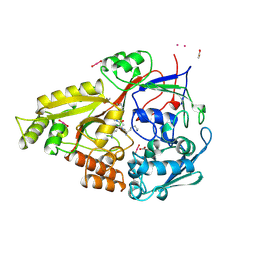 | | OLIGOPEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH TRI-LYSINE | | Descriptor: | ACETATE ION, OLIGO-PEPTIDE BINDING PROTEIN, TRIPEPTIDE LYS-LYS-LYS, ... | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1995-09-10 | | Release date: | 1996-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands.
Structure, 3, 1995
|
|
5CID
 
 | |
9C5S
 
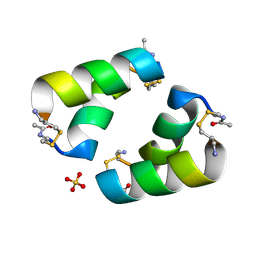 | | Disulfide-linked, antiparallel p53-derived peptide dimer (CV1) | | Descriptor: | Cellular tumor antigen p53, SULFATE ION | | Authors: | Vithanage, N, Kreitler, D.K, DiGiorno, M.C, Victorio, C.G, Sawyer, N, Outlaw, V.K. | | Deposit date: | 2024-06-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Characterization of Disulfide-Linked p53-Derived Peptide Dimers.
Res Sq, 2024
|
|
2YPP
 
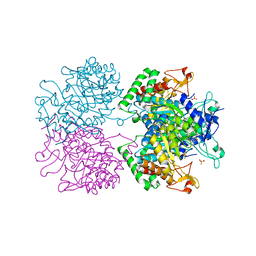 | | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase in complex with 3 tyrosine molecules | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Blackmore, N.J, Reichau, S, Jiao, W, Hutton, R.D, Baker, E.N, Jameson, G.B, Parker, E.J. | | Deposit date: | 2012-10-31 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three Sites and You are Out: Ternary Synergistic Allostery Controls Aromatic Aminoacid Biosynthesis in Mycobacterium Tuberculosis.
J.Mol.Biol., 425, 2013
|
|
8PV2
 
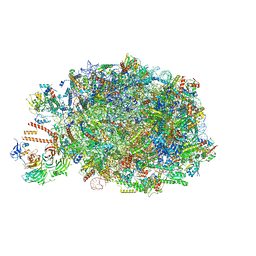 | | Chaetomium thermophilum pre-60S State 10 - pre-5S rotation with Ytm1-Erb1 | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV3
 
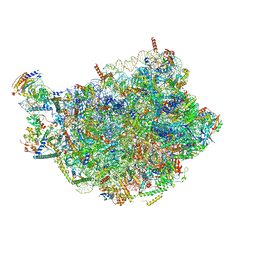 | | Chaetomium thermophilum pre-60S State 9 - pre-5S rotation - immature H68/H69 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV7
 
 | | Chaetomium thermophilum pre-60S State 1 - pre-5S rotation (Arx1/Nog2 state) - Composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVL
 
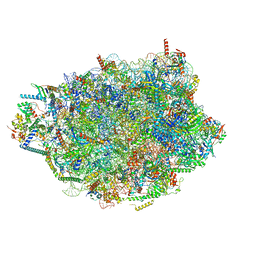 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV8
 
 | | Chaetomium thermophilum pre-60S State 4 - post-5S rotation with Rix1 complex without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
3A28
 
 | | Crystal structure of L-2,3-butanediol dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, L-2.3-butanediol dehydrogenase, MAGNESIUM ION, ... | | Authors: | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | Deposit date: | 2009-05-02 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chiral substrate recognition by two 2,3-butanediol dehydrogenases
Febs Lett., 584, 2010
|
|
1DAO
 
 | |
2YPO
 
 | | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with phenylalanine bound in only one site | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Blackmore, N.J, Reichau, S, Jiao, W, Hutton, R.D, Baker, E.N, Jameson, G.B, Parker, E.J. | | Deposit date: | 2012-10-31 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three Sites and You are Out: Ternary Synergistic Allostery Controls Aromatic Aminoacid Biosynthesis in Mycobacterium Tuberculosis.
J.Mol.Biol., 425, 2013
|
|
8OO0
 
 | | Chaetomium thermophilum Methionine Aminopeptidase 2 autoproteolysis product at the 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S0, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
