3PW2
 
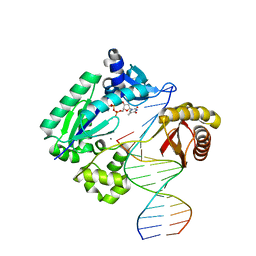 | |
3IEQ
 
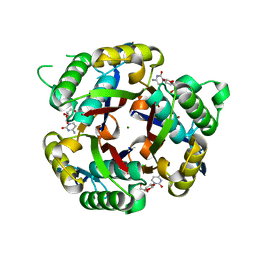 | |
4R8W
 
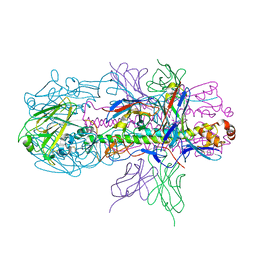 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a neutralizing antibody CT149 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of neutralizing antibody CT149, Hemagglutinin, ... | | Authors: | Wu, Y, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus
Nat Commun, 6, 2015
|
|
4RHK
 
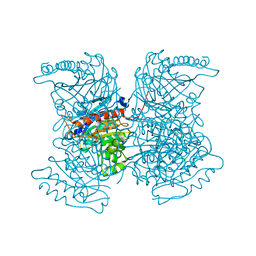 | |
3PHA
 
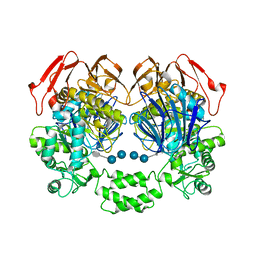 | | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose
To be Published
|
|
6BGD
 
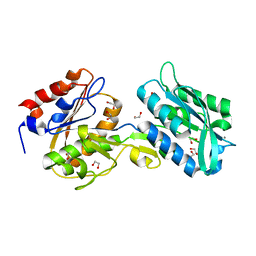 | | The crystal structure of the W145A variant of TpMglB-2 (Tp0684) with bound ligand | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucose/galactose-binding lipoprotein | | Authors: | Brautigam, C.A, Norgard, M.V, Deka, R.K. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structures of MglB-2 (TP0684), a topologically variant d-glucose-binding protein from Treponema pallidum, reveal a ligand-induced conformational change.
Protein Sci., 27, 2018
|
|
5QF8
 
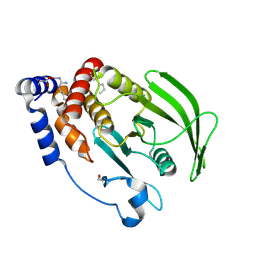 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000114a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7-hydroxy-2,2-dimethyl-2,3-dihydro-4H-1-benzopyran-4-one, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFT
 
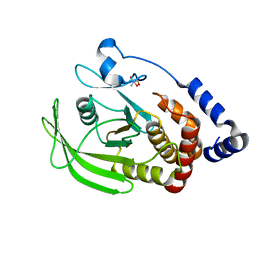 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMSOA000683b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-[(1H-pyrazol-1-yl)methyl]benzonitrile, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
3FZK
 
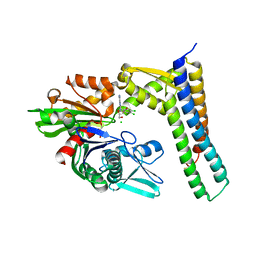 | | Crystal Structures of Hsc70/Bag1 in Complex with Small Molecule Inhibitors | | Descriptor: | (2R,3R,4S,5R)-2-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-5-(hydroxymethyl)oxolane-3,4-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG family molecular chaperone regulator 1, ... | | Authors: | Dokurno, P, Williamson, D.S, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design
J.Med.Chem., 52, 2009
|
|
5WMB
 
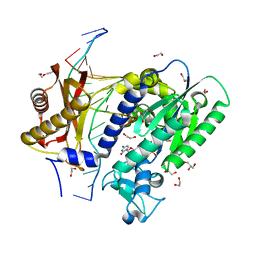 | | Structure of the 10S (-)-cis-BP-dG modified Rev1 ternary complex (the BP residue is disordered) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
5QG4
 
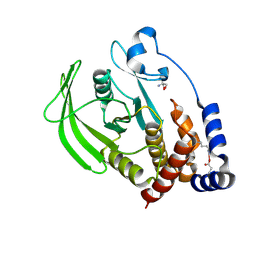 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000666a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, methyl (1S,3S,4R)-4-hydroxy-3-[(1S)-1-hydroxypropyl]-2-azabicyclo[2.2.2]octane-2-carboxylate | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
2XUS
 
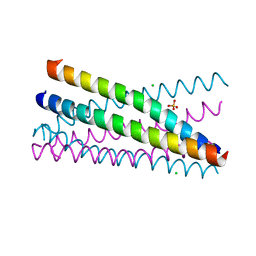 | | Crystal Structure of the BRMS1 N-terminal region | | Descriptor: | BREAST CANCER METASTASIS-SUPPRESSOR 1, CHLORIDE ION, SULFATE ION | | Authors: | Spinola-Amilibia, M, Rivera, J, Ortiz-Lombardia, M, Romero, A, Neira, J.L, Bravo, J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | The Structure of Brms1 Nuclear Export Signal and Snx6 Interacting Region Reveals a Hexamer Formed by Antiparallel Coiled Coils.
J.Mol.Biol., 411, 2011
|
|
6LFQ
 
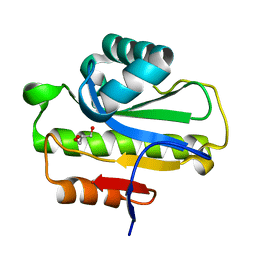 | | Crystal structure of Poa1p in apo form | | Descriptor: | ADP-ribose 1''-phosphate phosphatase, GLYCEROL | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
2XFG
 
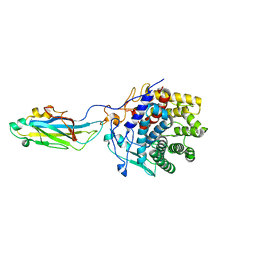 | | Reassembly and co-crystallization of a family 9 processive endoglucanase from separately expressed GH9 and CBM3c modules | | Descriptor: | CALCIUM ION, CHLORIDE ION, ENDOGLUCANASE 1 | | Authors: | Petkun, S, Lamed, R, Jindou, S, Burstein, T, Yaniv, O, Shoham, Y, Shimon, J.W.L, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Reassembly and Co-Crystallization of a Family 9 Processive Endoglucanase from its Component Parts: Structural and Functional Significance of Intermodular Linker
Peerj, 3, 2015
|
|
5QCQ
 
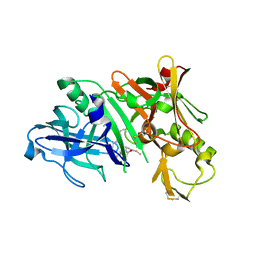 | | Crystal structure of BACE complex with BMC025 | | Descriptor: | (2R,4S,5S)-N-butyl-4-hydroxy-2,7-dimethyl-5-{[N-(4-methylpentanoyl)-L-methionyl]amino}octanamide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDD
 
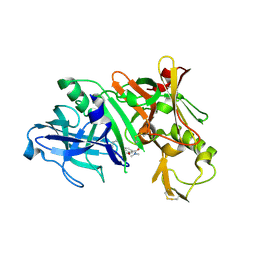 | | Crystal structure of BACE complex with BMC020 hydrolyzed | | Descriptor: | (10R,12S)-12-[(1R)-1,2-dihydroxyethyl]-N,N,10-trimethyl-14-oxo-2-oxa-13-azabicyclo[13.3.1]nonadeca-1(19),15,17-triene-17-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5WU3
 
 | | Crystal structure of human Tut1 bound with MgUTP, form II | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
5QFD
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000505a | | Descriptor: | (6R)-5,6-dihydro-1H-2,6-methano-1lambda~6~-1lambda~6~,2,5-benzothiadiazocine-1,1,4(3H)-trione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFU
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000487a | | Descriptor: | (1R,4R,5R,6R)-4-methoxy-2-(methylsulfonyl)-2-azabicyclo[3.3.1]nonan-6-ol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.613 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5CZX
 
 | | Crystal structure of Notch3 NRR in complex with 20358 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 20358 Fab heavy chain, ... | | Authors: | Hu, T, Fryer, C, Chopra, R, Clark, K. | | Deposit date: | 2015-08-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies.
Oncogene, 35, 2016
|
|
2XEC
 
 | | Nocardia farcinica maleate cis-trans isomerase bound to TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, PUTATIVE MALEATE ISOMERASE | | Authors: | Fisch, F, Martinez-Fleites, C, Baudendistel, N, Hauer, B, Turkenburg, J.P, Hart, S, Bruce, N.C, Grogan, G. | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Covalent Succinylcysteine-Like Intermediate in the Enzyme-Catalyzed Transformation of Maleate to Fumarate by Maleate Isomerase.
J.Am.Chem.Soc., 132, 2010
|
|
4A4X
 
 | | NEK2-EDE bound to CCT248662 | | Descriptor: | 4-(2-AMINO-5-{4-[(DIMETHYLAMINO)METHYL]THIOPHEN-2-YL}PYRIDIN-3-YL)-2-{(1R)-1-[2-(TRIFLUOROMETHYL)PHENYL]ETHOXY}BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2011-10-20 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of Potent and Selective Hybrid Inhibitors of the Mitotic Kinase Nek2: Structure-Activity Relationship, Structural Biology, and Cellular Activity.
J.Med.Chem., 55, 2012
|
|
5W9E
 
 | | Toxoplasma Gondii CDPK1 in complex with inhibitor GXJ-186 | | Descriptor: | 1-tert-butyl-3-[(3-chlorophenyl)sulfanyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1 | | Authors: | El Bakkouri, M, Lovato, D, Loppnau, P, Lin, Y.H, Rutaganaria, F, Lopez, M.S, Shokat, L, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Sibley, D, Hui, R, Walker, J.R. | | Deposit date: | 2017-06-23 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Toxoplasma Gondii CDPK1 in complex with inhibitor GXJ-186
To be published
|
|
5QD3
 
 | | Crystal structure of BACE complex with BMC010 | | Descriptor: | (10R,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5TA7
 
 | | Crystal structure of BuGH117Bwt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
