8PK7
 
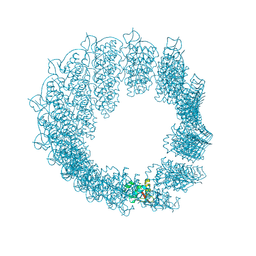 | | Helical reconstruction of CHIKV nsP3 helical scaffolds | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Reguera, J, Hons, M, Zimberger, C, Ptchelkine, D, Jones, R, Desfosses, A. | | Deposit date: | 2023-06-25 | | Release date: | 2024-08-14 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Alphavirus nsP3 organizes into tubular scaffolds essential for infection and the cytoplasmic granule architecture.
Nat Commun, 15, 2024
|
|
8PHZ
 
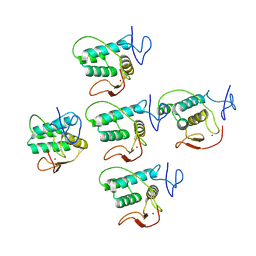 | | Helical reconstruction of CHIKV nsP3 helical scaffolds | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Reguera, J, Hons, M, Zimberger, C, Ptchelkine, D, Jones, R, Desfosses, A. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-14 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Alphavirus nsP3 organizes into tubular scaffolds essential for infection and the cytoplasmic granule architecture.
Nat Commun, 15, 2024
|
|
5HIC
 
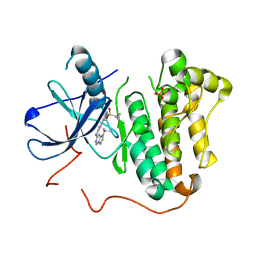 | | EGFR kinase domain mutant "TMLR" with a imidazopyridinyl-aminopyrimidine inhibitor | | Descriptor: | Epidermal growth factor receptor, N-{2-[1-(cyclopropylsulfonyl)-1H-pyrazol-4-yl]pyrimidin-4-yl}-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-amine, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
9FL1
 
 | |
9FOB
 
 | |
5HIB
 
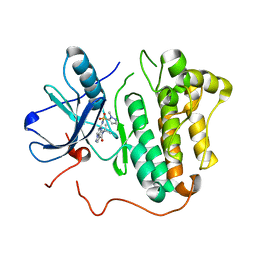 | | EGFR kinase domain mutant "TMLR" with a pyrazolopyrimidine inhibitor | | Descriptor: | Epidermal growth factor receptor, N-tert-butyl-5-{[(1-methyl-1H-pyrazol-5-yl)sulfonyl]amino}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
7AGH
 
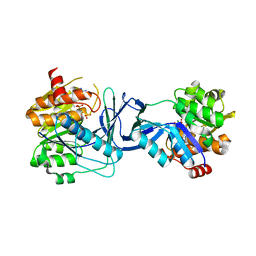 | |
7AG7
 
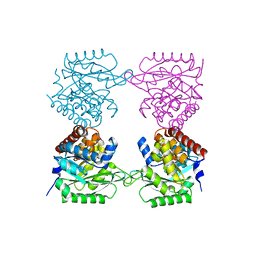 | |
7AG4
 
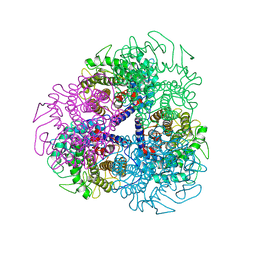 | |
7AG1
 
 | |
7AG6
 
 | |
7AGK
 
 | | Crystal structure of E. coli SF kinase (YihV) in complex with product sulfofructose phosphate (SFP) | | Descriptor: | Sulfofructose kinase, [(2~{S},3~{S},4~{S},5~{R})-3,4,5-tris(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
5F6Q
 
 | | Crystal Structure of Metallothiol Transferase from Bacillus anthracis str. Ames | | Descriptor: | CHLORIDE ION, GLYCEROL, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Metallothiol Transferase from Bacillus anthracis str. Ames
To Be Published
|
|
9G1H
 
 | | Fragment screening of FosAKP, room-temperature structure in complex with fragment F2X-entry H01 | | Descriptor: | Fosfomycin resistance protein, MANGANESE (II) ION, ~{N},~{N}-diethyl-2-(4-nitrophenoxy)ethanamine | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Room temperature X-ray fragment screening with serial crystallography
To Be Published
|
|
9G1A
 
 | | Fragment screening of FosAKP, room-temperature structure, ground state | | Descriptor: | Fosfomycin resistance protein, MANGANESE (II) ION | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Room temperature X-ray fragment screening with serial crystallography
To Be Published
|
|
9G1G
 
 | | Fragment screening of FosAKP, room-temperature structure in complex with fragment F2X-entry G08 | | Descriptor: | Fosfomycin resistance protein, MANGANESE (II) ION, N-ethyl-2-{[5-(propan-2-yl)-1,3,4-oxadiazol-2-yl]sulfanyl}acetamide | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Room temperature X-ray fragment screening with serial crystallography
To Be Published
|
|
6ZA9
 
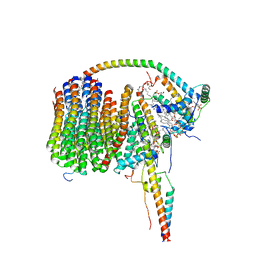 | | Fo domain of Ovine ATP synthase | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit C1, mitochondrial, ... | | Authors: | Pinke, G, Zhou, L, Sazanov, L.A. | | Deposit date: | 2020-06-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Cryo-EM structure of the entire mammalian F-type ATP synthase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1JUN
 
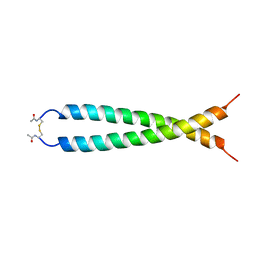 | |
1FXL
 
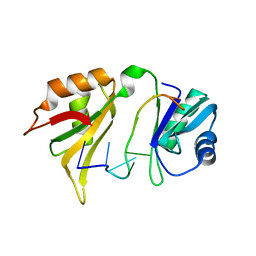 | |
6U3F
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Panton-Valentine Leucocidin F, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U2S
 
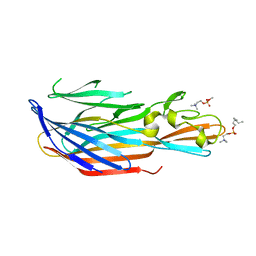 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Bi-component leukocidin LukED subunit D, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U3T
 
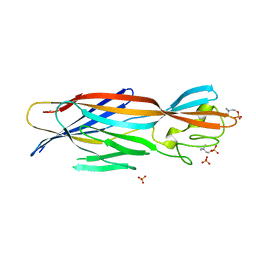 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U4P
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-26 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U3Y
 
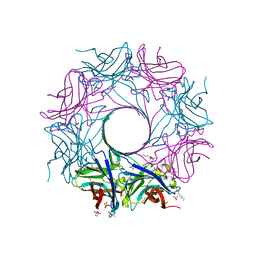 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | ACETATE ION, Gamma-hemolysin subunit A, Panton-Valentine Leucocidin F, ... | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U49
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
