1LVE
 
 | |
1RAW
 
 | |
1NFN
 
 | | APOLIPOPROTEIN E3 (APOE3) | | Descriptor: | APOLIPOPROTEIN E3 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1ANU
 
 | | COHESIN-2 DOMAIN OF THE CELLULOSOME FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | COHESIN-2 | | Authors: | Shimon, L.J.W, Yaron, S, Shoham, Y, Lamed, R, Morag, E, Bayer, E.A, Frolow, F. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A cohesin domain from Clostridium thermocellum: the crystal structure provides new insights into cellulosome assembly.
Structure, 5, 1997
|
|
1JHG
 
 | | TRP REPRESSOR MUTANT V58I | | Descriptor: | PHOSPHATE ION, TRP OPERON REPRESSOR, TRYPTOPHAN | | Authors: | Lawson, C.L. | | Deposit date: | 1996-07-19 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An atomic view of the L-tryptophan binding site of trp repressor.
Nat.Struct.Biol., 3, 1996
|
|
1QCM
 
 | | AMYLOID BETA PEPTIDE (25-35), NMR, 20 STRUCTURES | | Descriptor: | AMYLOID BETA PEPTIDE | | Authors: | Kohno, T, Kobayashi, K, Maeda, T, Sato, K, Takashima, A. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structures of the amyloid beta peptide (25-35) in membrane-mimicking environment.
Biochemistry, 35, 1996
|
|
1TOC
 
 | |
1TXA
 
 | | SOLUTION NMR STRUCTURE OF TOXIN B, A LONG NEUROTOXIN FROM THE VENOM OF THE KING COBRA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TOXIN B | | Authors: | Peng, S.-S, Kumar, T.K.S, Jayaraman, G, Chang, C.-C, Yu, C. | | Deposit date: | 1996-07-20 | | Release date: | 1997-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of toxin b, a long neurotoxin from the venom of the king cobra (Ophiophagus hannah).
J.Biol.Chem., 272, 1997
|
|
1KSI
 
 | | CRYSTAL STRUCTURE OF A EUKARYOTIC (PEA SEEDLING) COPPER-CONTAINING AMINE OXIDASE AT 2.2A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Wilce, M.C.J, Kumar, V, Freeman, H.C, Guss, J.M. | | Deposit date: | 1996-07-20 | | Release date: | 1997-12-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 A resolution.
Structure, 4, 1996
|
|
1XVA
 
 | | METHYLTRANSFERASE | | Descriptor: | ACETATE ION, GLYCINE N-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Fu, Z, Hu, Y, Konishi, K, Takata, Y, Ogawa, H, Gomi, T, Fujioka, M, Takusagawa, F. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycine N-methyltransferase from rat liver.
Biochemistry, 35, 1996
|
|
1TXB
 
 | | SOLUTION NMR STRUCTURE OF TOXIN B, A LONG NEUROTOXIN FROM THE VENOM OF THE KING COBRA, 10 STRUCTURES | | Descriptor: | TOXIN B | | Authors: | Peng, S.-S, Kumar, T.K.S, Jayaraman, G, Chang, C.-C, Yu, C. | | Deposit date: | 1996-07-20 | | Release date: | 1997-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of toxin b, a long neurotoxin from the venom of the king cobra (Ophiophagus hannah).
J.Biol.Chem., 272, 1997
|
|
1LVY
 
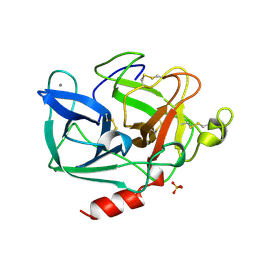 | | PORCINE ELASTASE | | Descriptor: | CALCIUM ION, ELASTASE, SULFATE ION | | Authors: | Schiltz, M, Prange, T. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | High-pressure krypton gas and statistical heavy-atom refinement: a successful combination of tools for macromolecular structure determination.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1EDK
 
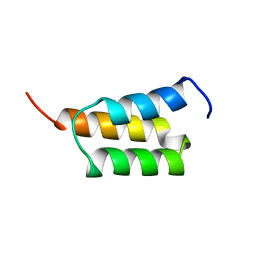 | |
1EDL
 
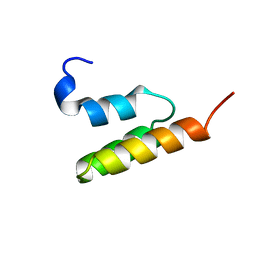 | |
1KBT
 
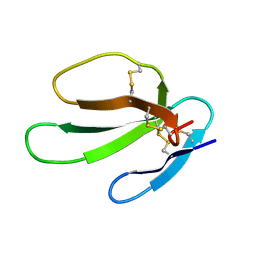 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 12 STRUCTURES | | Descriptor: | CTX IV | | Authors: | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
1KBS
 
 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 1 STRUCTURE | | Descriptor: | CTX IV | | Authors: | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
278D
 
 | |
276D
 
 | |
1NSY
 
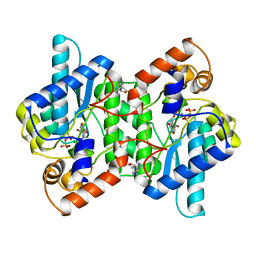 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rizzi, M, Nessi, C, Bolognesi, M, Galizzi, A, Coda, A. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Bacillus subtilis.
EMBO J., 15, 1996
|
|
288D
 
 | |
277D
 
 | |
1WPO
 
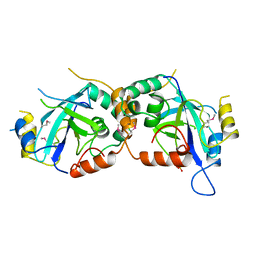 | | HYDROLYTIC ENZYME HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | HUMAN CYTOMEGALOVIRUS PROTEASE, SULFATE ION | | Authors: | Tong, L. | | Deposit date: | 1996-07-23 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease.
Nature, 383, 1996
|
|
1NAW
 
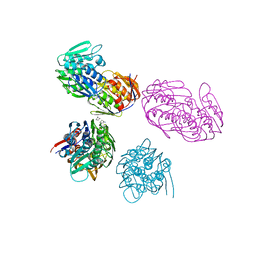 | | ENOLPYRUVYL TRANSFERASE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | Authors: | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | Deposit date: | 1996-07-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
1UGD
 
 | |
3MHT
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH UNMODIFIED DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), PROTEIN (HHAI METHYLTRANSFERASE (E.C.2.1.1.73)), ... | | Authors: | O'Gara, M, Klimasauskas, S, Roberts, R.J, Cheng, X. | | Deposit date: | 1996-07-24 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes.
J.Mol.Biol., 261, 1996
|
|
