4TVU
 
 | | Crystal structure of trehalose synthase from Deinococcus radiodurans reveals a closed conformation for catalysis of the intramolecular isomerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wang, Y.L, Chow, S.Y, Lin, Y.T, Liaw, S.H. | | Deposit date: | 2014-06-28 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5L8D
 
 | |
5HXV
 
 | |
5DMQ
 
 | |
9HC0
 
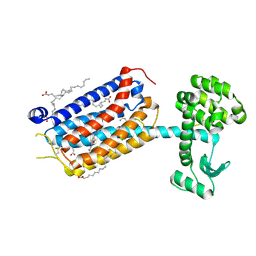 | | Dark structure of the human metabotropic glutamate receptor 5 transmembrane domain bound to photoswitchable ligand alloswitch-1 | | Descriptor: | 2-chloranyl-~{N}-[2-methoxy-4-[(~{E})-pyridin-2-yldiazenyl]phenyl]benzamide, Metabotropic glutamate receptor 5,Endolysin, OLEIC ACID | | Authors: | Kondo, Y, Hatton, C, Cheng, R, Trabuco, M, Glover, H, Bertrand, Q, Stierli, F, Seidel, H.P, Mason, T, Sarma, S, Tellkamp, F, Kepa, M, Dworkowski, F, Mehrabi, P, Hennig, M, Standfuss, J. | | Deposit date: | 2024-11-08 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Apo-state structure of the metabotropic glutamate receptor 5 transmembrane domain obtained using a photoswitchable ligand.
Protein Sci., 34, 2025
|
|
5ZH5
 
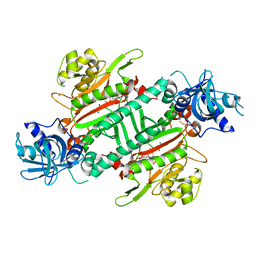 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-2 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6S)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
5ZIX
 
 | |
9HC3
 
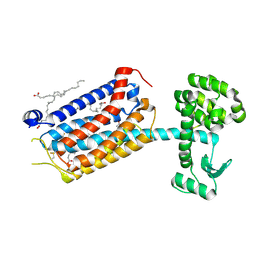 | | Apo-state structure of the human metabotropic glutamate receptor 5 transmembrane domain freeze-trapped after light activation of photoswitchable ligand alloswitch-1 | | Descriptor: | Metabotropic glutamate receptor 5,Endolysin, OLEIC ACID | | Authors: | Kondo, Y, Hatton, C, Cheng, R, Trabuco, M, Glover, H, Bertrand, Q, Stierli, F, Seidel, H.P, Mason, T, Sarma, S, Tellkamp, F, Kepa, M, Dworkowski, F, Mehrabi, P, Hennig, M, Standfuss, J. | | Deposit date: | 2024-11-08 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Apo-state structure of the metabotropic glutamate receptor 5 transmembrane domain obtained using a photoswitchable ligand.
Protein Sci., 34, 2025
|
|
4TZO
 
 | |
8TC9
 
 | | Human asparaginyl-tRNA synthetase bound to OSM-S-106 | | Descriptor: | Asparagine--tRNA ligase, cytoplasmic, GLYCEROL, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
5LW3
 
 | | Azotobacter vinelandii Mannuronan C-5 epimerase AlgE6 A-module | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rothweiler, U. | | Deposit date: | 2016-09-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Azotobacter vinelandii Mannuronan C-5 epimerase AlgE6 A-module
To Be Published
|
|
9GYP
 
 | | Crystal structure of human Histidine Triad Nucleotide-Binding Protein 1 in complex with KV24 | | Descriptor: | Histidine triad nucleotide-binding protein 1, SULFATE ION, ~{N}-oxidanyl-4-(5~{H}-pyrrolo[1,2-a]quinoxalin-4-yl)benzamide | | Authors: | Dolot, R.M, Lechner, S, Sethiya, J.P, Wagner, C.R, Bracher, F, Kuster, B. | | Deposit date: | 2024-10-02 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Serendipitous and Systematic Chemoproteomic Discovery of MBLAC2, HINT1, and NME1-4 Inhibitors from Histone Deacetylase-Targeting Pharmacophores.
Acs Chem.Biol., 20, 2025
|
|
7T1W
 
 | |
6SHZ
 
 | | p53 cancer mutant Y220C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
6SD9
 
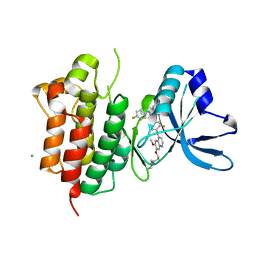 | | Crystal structure of wild-type cMET bound by foretinib | | Descriptor: | CHLORIDE ION, Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDC
 
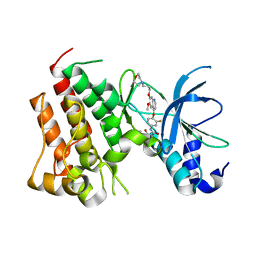 | | Crystal structure of D1228V cMET bound by foretinib | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6IBS
 
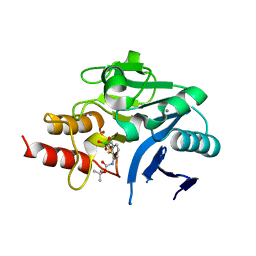 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 6 | | Descriptor: | CALCIUM ION, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6FGG
 
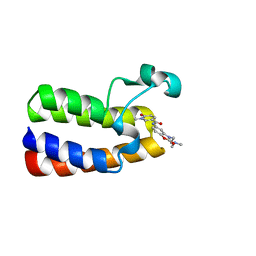 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 5 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[3-[2-(dimethylamino)ethyl]-2-oxidanylidene-1,3-benzoxazol-5-yl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
5HWL
 
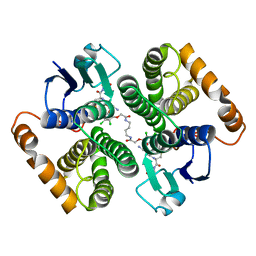 | | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 2, N,N'-(butane-1,4-diyl)bis{2-[2,3-dichloro-4-(2-methylidenebutanoyl)phenoxy]acetamide} | | Authors: | Zhang, X, Wei, J, Wu, S, Zhang, H.P, Luo, M, Yang, X.L, Liao, F, Wang, D.Q. | | Deposit date: | 2016-01-29 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form
To Be Published
|
|
3DN7
 
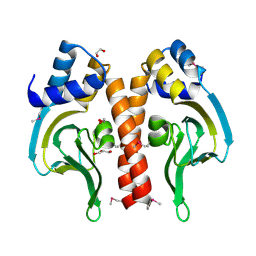 | | Cyclic nucleotide binding regulatory protein from Cytophaga hutchinsonii. | | Descriptor: | 1,2-ETHANEDIOL, Cyclic nucleotide binding regulatory protein, GLYCEROL | | Authors: | Osipiuk, J, Maltseva, N, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of cyclic nucleotide binding regulatory protein from Cytophaga hutchinsonii.
To be Published
|
|
8YIY
 
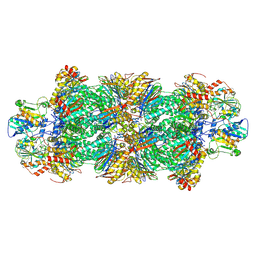 | | Cryo-EM structure of human proteasome assembly intermediate preholo-1 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Han, Y, Han, Q, Tang, Q, Zhang, Y, Liu, K. | | Deposit date: | 2024-02-29 | | Release date: | 2025-01-15 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis for the stepwise and faithful maturation of the 20 S proteasome.
Sci Adv, 11, 2025
|
|
5JO1
 
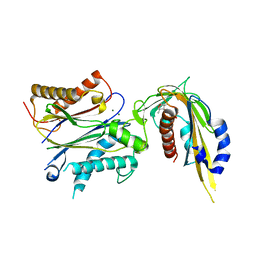 | | Crystal structure of phaseic acid-bound abscisic acid receptor PYL3 in complex with type 2C protein phosphatase HAB1 | | Descriptor: | (3S,4E)-5-[(1R,5R,8S)-8-hydroxy-1,5-dimethyl-3-oxo-6-oxabicyclo[3.2.1]octan-8-yl]-3-methylpent-4-enoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION, ... | | Authors: | Weng, J.K, Noel, J.P. | | Deposit date: | 2016-05-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-evolution of Hormone Metabolism and Signaling Networks Expands Plant Adaptive Plasticity.
Cell, 166, 2016
|
|
5ZBK
 
 | |
6DX5
 
 | | Crystal structure of the viral OTU domain protease from Farallon virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, RNA-dependent RNA polymerase | | Authors: | Beldon, B.S, Dzimianski, J.V, Daczkowski, C.M, Goodwin, O.Y, Pegan, S.D. | | Deposit date: | 2018-06-28 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Probing the impact of nairovirus genomic diversity on viral ovarian tumor domain protease (vOTU) structure and deubiquitinase activity.
PLoS Pathog., 15, 2019
|
|
6DXQ
 
 | | Crystal structure of the LigJ Hydratase product complex with 4-carboxy-4-hydroxy-2-oxoadipate | | Descriptor: | (2S)-2-hydroxy-4-oxobutane-1,2,4-tricarboxylic acid, 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M. | | Deposit date: | 2018-06-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
