7Y60
 
 | | Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5L
 
 | | Crystal structure of human CAF-1 core complex in spacegroup C2 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8AF2
 
 | | Human Sterol Carrier Protein with unnatural amino acid 2,2'-bipyridine alanine incorporated at position 111 | | Descriptor: | COPPER (II) ION, Enoyl-CoA hydratase 2, FRAGMENT OF TRITON X-100, ... | | Authors: | Richardson, J.M, Klemencic, E, Jarvis, A.G. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Using BpyAla to generate copper artificial metalloenzymes: a catalytic and structural study.
Catalysis Science And Technology, 14, 2024
|
|
8AF3
 
 | | Sterol carrier protein Artifical metalloenzyme incorporating Q111C mutation coupled to 2,2'-bipyridine | | Descriptor: | COPPER (II) ION, Enoyl-CoA hydratase 2, FRAGMENT OF TRITON X-100, ... | | Authors: | Richardson, J.M, Klemencic, E, Jarvis, A.G. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Using BpyAla to generate copper artificial metalloenzymes: a catalytic and structural study.
Catalysis Science And Technology, 14, 2024
|
|
8BQE
 
 | | In situ structure of the Caulobacter crescentus S-layer | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose, CALCIUM ION, S-layer protein rsaA | | Authors: | von Kuegelgen, A, Bharat, T. | | Deposit date: | 2022-11-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0.
Elife, 11, 2022
|
|
8BSH
 
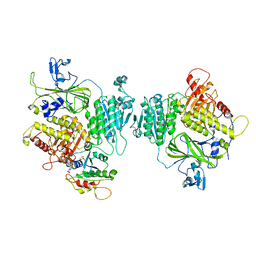 | | COPII inner coat | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein transport protein SEC23, ... | | Authors: | Zanetti, G, Pyle, E.W. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0.
Elife, 11, 2022
|
|
7R3K
 
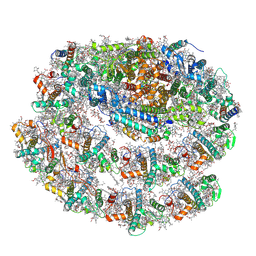 | | Chlamydomonas reinhardtii TSP9 mutant small Photosystem I complex | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R)-beta,beta-caroten-3-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Klaiman, D, Schwartz, T, Nelson, N. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structure of Photosystem I Supercomplex Isolated from a Chlamydomonas reinhardtii Cytochrome b6f Temperature-Sensitive Mutant.
Biomolecules, 13, 2023
|
|
4UB8
 
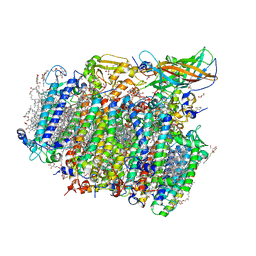 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
4UB6
 
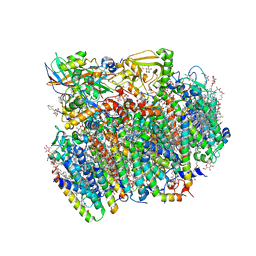 | | Native structure of photosystem II (dataset-1) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
8OI7
 
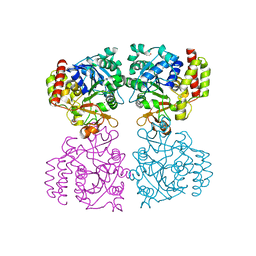 | | Trichomonas vaginalis riboside hydrolase | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Inosine-uridine preferring nucleoside hydrolase family protein, ... | | Authors: | Patrone, M, Stockman, B.J, Degano, M. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A riboside hydrolase that salvages both nucleobases and nicotinamide in the auxotrophic parasite Trichomonas vaginalis.
J.Biol.Chem., 299, 2023
|
|
8OI9
 
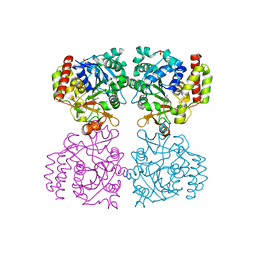 | | Trichomonas vaginalis riboside hydrolase in complex with 5-methyluridine | | Descriptor: | 5-methyluridine, CALCIUM ION, Inosine-uridine preferring nucleoside hydrolase family protein, ... | | Authors: | Patrone, M, Stockman, B.J, Degano, M. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A riboside hydrolase that salvages both nucleobases and nicotinamide in the auxotrophic parasite Trichomonas vaginalis.
J.Biol.Chem., 299, 2023
|
|
8OIA
 
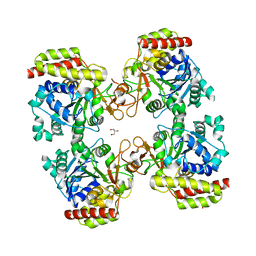 | | Trichomonas vaginalis riboside hydrolase in complex with D-ribose | | Descriptor: | CALCIUM ION, GLYCEROL, Inosine-uridine preferring nucleoside hydrolase family protein, ... | | Authors: | Patrone, M, Stockman, B.J, Degano, M. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A riboside hydrolase that salvages both nucleobases and nicotinamide in the auxotrophic parasite Trichomonas vaginalis.
J.Biol.Chem., 299, 2023
|
|
8OIC
 
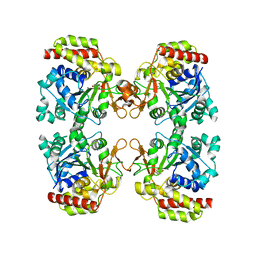 | | Trichomonas vaginalis riboside hydrolase (His-tagged) | | Descriptor: | BICINE, CALCIUM ION, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Patrone, M, Stockman, B.J, Degano, M. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A riboside hydrolase that salvages both nucleobases and nicotinamide in the auxotrophic parasite Trichomonas vaginalis.
J.Biol.Chem., 299, 2023
|
|
8OIB
 
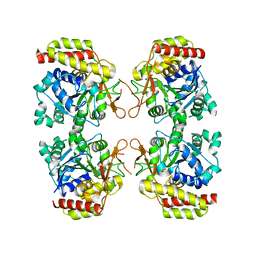 | |
8K1L
 
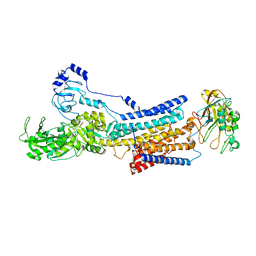 | | Cryo-EM structure of Na+,K+-ATPase alpha2 from Artemia salina in cation-free E2P form | | Descriptor: | Na+,K+-ATPase alpha2KK, Na+,K+-ATPase beta2, TETRAFLUOROALUMINATE ION | | Authors: | Abe, K, Artigas, P. | | Deposit date: | 2023-07-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | A Na pump with reduced stoichiometry is up-regulated by brine shrimp in extreme salinities.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8IQG
 
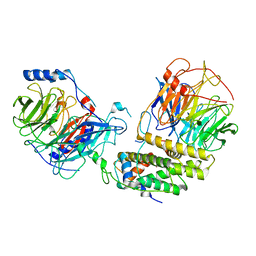 | | Cryo-EM structure of the monomeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQF
 
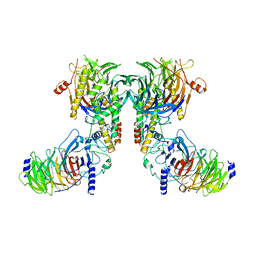 | | Cryo-EM structure of the dimeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8I0E
 
 | | Sb3GT1 complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, H.T, Wang, Z.L, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
8HZZ
 
 | | GuApiGT (UGT79B74) | | Descriptor: | SULFATE ION, apiosyltransferase | | Authors: | Wang, H.T, Wang, Z.L, Li, F.D, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
8I0D
 
 | | Sb3GT1 375S/Q377H mutant complex with UDP-Glc | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Wang, H.T, Wang, Z.L, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
8J6T
 
 | | Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8J6S
 
 | | Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8JAT
 
 | |
8QTQ
 
 | | Thermostable WW domain | | Descriptor: | WW domain | | Authors: | Kovermann, M, Thomas, F. | | Deposit date: | 2023-10-13 | | Release date: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Thermostable WW-Domain Scaffold to Design Functional beta-Sheet Miniproteins.
J.Am.Chem.Soc., 2024
|
|
7V6P
 
 | | Crystal structure of human sNASP TPR domain | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Liu, C.P, Xu, R.M. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Distinct histone H3-H4 binding modes of sNASP reveal the basis for cooperation and competition of histone chaperones.
Genes Dev., 35, 2021
|
|
