1E4P
 
 | |
1E4Q
 
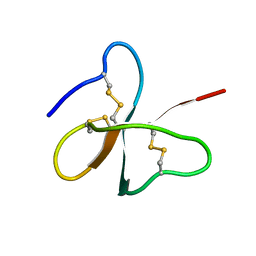 | | Solution structure of the human defensin hBD-2 | | Descriptor: | BETA-DEFENSIN 2 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
1E4R
 
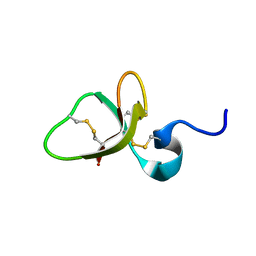 | | Solution structure of the mouse defensin mBD-8 | | Descriptor: | Beta-defensin 8 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1E4S
 
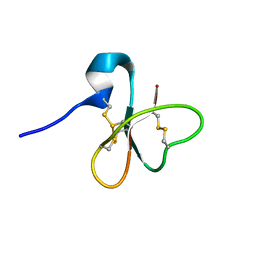 | | Solution structure of the human defensin hBD-1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
1E4T
 
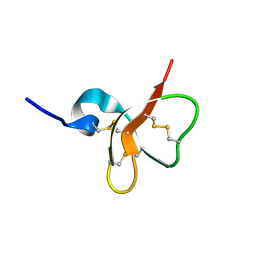 | | Solution structure of the mouse defensin mBD-7 | | Descriptor: | Beta-defensin 7 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1E4U
 
 | | N-terminal RING finger domain of human NOT-4 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR NOT4, ZINC ION | | Authors: | Hanzawa, H, De Ruwe, M.J, Albert, T.K, Van Der Vliet, P.C, Timmers, H.T, Boelens, R. | | Deposit date: | 2000-07-12 | | Release date: | 2001-03-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the C4C4 Ring Finger of Human not4 Reveals Features Distinct from Those of C3Hc4 Ring Fingers
J.Biol.Chem., 276, 2001
|
|
1E4V
 
 | |
1E4W
 
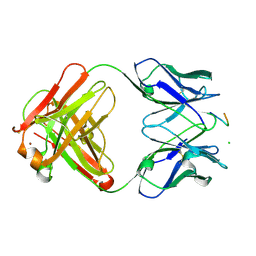 | | crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment | | Descriptor: | CHLORIDE ION, CYCLIC PEPTIDE, NICKEL (II) ION, ... | | Authors: | Hahn, M, Winkler, D, Misselwitz, R, Wessner, H, Welfle, K, Zahn, G, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-Reactive Binding of Cyclic Peptides to an Anti-Tgf Alpha Antibody Fab Fragment: An X-Ray Structural and Thermodynamic Analysis
J.Mol.Biol., 314, 2001
|
|
1E4X
 
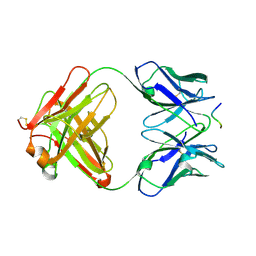 | | crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment | | Descriptor: | CYCLIC PEPTIDE, TAB2 | | Authors: | Hahn, M, Winkler, D, Misselwitz, R, Wessner, H, Welfle, K, Zahn, G, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Reactive Binding of Cyclic Peptides to an Anti-Tgf Alpha Antibody Fab Fragment: An X-Ray Structural and Thermodynamic Analysis
J.Mol.Biol., 314, 2001
|
|
1E4Y
 
 | |
1E50
 
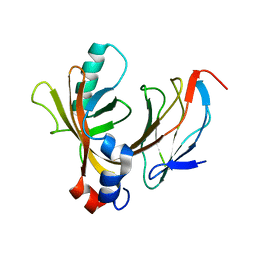 | | AML1/CBFbeta complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT, CORE-BINDING FACTOR CBF-BETA | | Authors: | Warren, A.J, Bravo, J, Williams, R.L, Rabbits, T.H. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Heterodimeric Interaction between the Acute Leukaemia-Associated Transcription Factors Aml1 and Cbfbeta
Embo J., 19, 2000
|
|
1E51
 
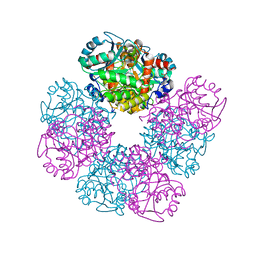 | | Crystal structure of native human erythrocyte 5-aminolaevulinic acid dehydratase | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, SULFATE ION, ... | | Authors: | Mills-Davies, N.L, Thompson, D, Cooper, J.B, Shoolingin-Jordan, P.M. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of Human Ala-Dehydratase
To be Published
|
|
1E52
 
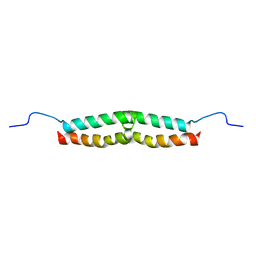 | | Solution structure of Escherichia coli UvrB C-terminal domain | | Descriptor: | EXCINUCLEASE ABC SUBUNIT | | Authors: | Alexandrovich, A.A, Kelly, G.G, Frenkiel, T.A, Moolenaar, G.F, Goosen, N.N, Sanderson, M.R, Lane, A.N. | | Deposit date: | 2000-07-14 | | Release date: | 2001-07-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Hydrodynamics and Thermodynamics of the Uvrb C-Terminal Domain.
J.Biomol.Struct.Dyn., 19, 2001
|
|
1E54
 
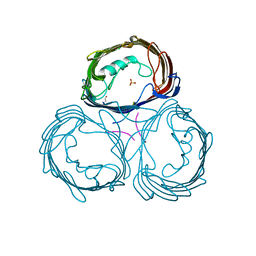 | | Anion-selective porin from Comamonas acidovorans | | Descriptor: | CALCIUM ION, OMP32, OUTER MEMBRANE PORIN PROTEIN 32, ... | | Authors: | Zeth, K, Diederichs, K, Welte, W, Engelhardt, H. | | Deposit date: | 2000-07-17 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Omp32, the Anion-Selective Porin from Comamonas Acidovorans, in Complex with a Periplasmic Peptideat 2.1 A Resolution
Structure, 8, 2000
|
|
1E55
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the competitive inhibitor dhurrin | | Descriptor: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, BETA-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of substrate (aglycone) specificity in beta-glucosidases is revealed by crystal structures of mutant maize beta-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes.
Proc. Natl. Acad. Sci. U.S.A., 97, 2000
|
|
1E56
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the natural substrate DIMBOA-beta-D-glucoside | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase- Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E57
 
 | |
1E58
 
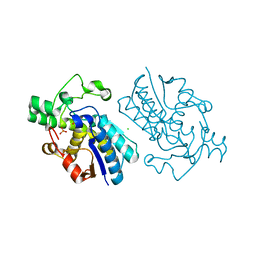 | | E.coli cofactor-dependent phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCERATE MUTASE, SULFATE ION | | Authors: | Bond, C.S, Hunter, W.N. | | Deposit date: | 2000-07-19 | | Release date: | 2001-03-20 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High Resolution Structure of the Phosphohistidine-Activated Form of Escherichia Coli Cofactor-Dependent Phosphoglycerate Mutase.
J.Biol.Chem., 276, 2001
|
|
1E59
 
 | |
1E5A
 
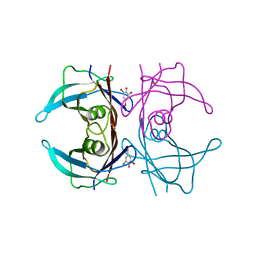 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | 2,4,6-TRIBROMOPHENOL, TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E5B
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-07-24 | | Release date: | 2001-05-25 | | Last modified: | 2018-10-24 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
1E5C
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-07-24 | | Release date: | 2001-05-25 | | Last modified: | 2018-10-24 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
1E5D
 
 | | RUBREDOXIN OXYGEN:OXIDOREDUCTASE (ROO) FROM ANAEROBE DESULFOVIBRIO GIGAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, MU-OXO-DIIRON, OXYGEN MOLECULE, ... | | Authors: | Frazao, C, Silva, G, Gomes, C.M, Matias, P, Coelho, R, Sieker, L, Macedo, S, Liu, M.Y, Oliveira, S, Teixeira, M, Xavier, A.V, Rodrigues-Pousada, C, Carrondo, M.A, Le Gall, J. | | Deposit date: | 2000-07-24 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Dioxygen Reduction Enzyme from Desulfovibrio Gigas
Nat.Struct.Biol., 7, 2000
|
|
1E5E
 
 | | METHIONINE GAMMA-LYASE (MGL) FROM TRICHOMONAS VAGINALIS IN COMPLEX WITH PROPARGYLGLYCINE | | Descriptor: | GLYCEROL, METHIONINE GAMMA-LYASE, N-(HYDROXY{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)NORVALINE, ... | | Authors: | Goodall, G, Mottram, J.C, Coombs, G.H, Lapthorn, A.J. | | Deposit date: | 2000-07-25 | | Release date: | 2001-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Structure and Proposed Catalytic Mechanism of Methionine Gamma-Lyase
To be Published
|
|
1E5F
 
 | | METHIONINE GAMMA-LYASE (MGL) FROM TRICHOMONAS VAGINALIS | | Descriptor: | GLYCEROL, METHIONINE GAMMA-LYASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Goodall, G, Mottram, J.C, Coombs, G.H, Lapthorn, A.J. | | Deposit date: | 2000-07-25 | | Release date: | 2001-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Structure and Proposed Catalytic Mechanism of Methionine Gamma-Lyase
To be Published
|
|
