7EWC
 
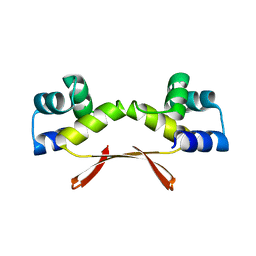 | | Mycobacterium tuberculosis HigA2 (Form I) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | Descriptor: | HTH cro/C1-type domain-containing protein | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSY
 
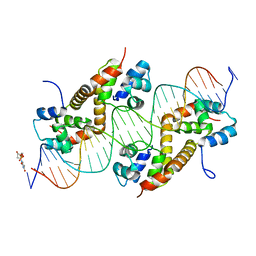 | | Pseudomonas aeruginosa antitoxin HigA with higBA promoter | | Descriptor: | DNA (28-MER), DNA (29-MER), HTH cro/C1-type domain-containing protein, ... | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
3FYA
 
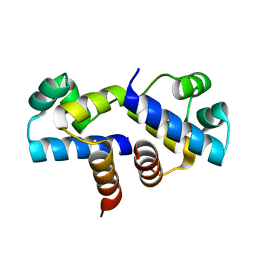 | | Crystal Structure of an R35A mutant of the Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | Regulatory protein | | Authors: | Ball, N.J, McGeehan, J.E, Thresh, S.J, Streeter, S.D, Kneale, G.G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the restriction-modification controller protein C.Esp1396I.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3G5G
 
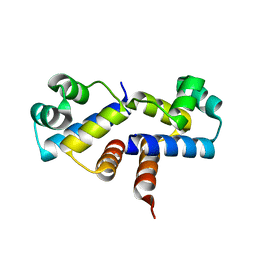 | | Crystal Structure of the Wild-Type Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | Regulatory protein | | Authors: | Ball, N.J, McGeehan, J.E, Thresh, S.J, Streeter, S.D, Kneale, G.G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the restriction-modification controller protein C.Esp1396I.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6H49
 
 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | Descriptor: | Orf20, SULFATE ION | | Authors: | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
6RNX
 
 | | Crystal structure of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6RMQ
 
 | | Crystal structure of a selenomethionine-substituted A70M I84M mutant of the essential repressor DdrO from radiation resistant-Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
2R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
6RNZ
 
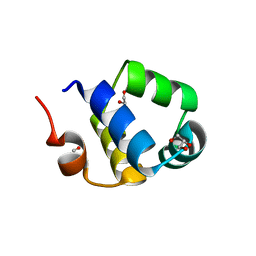 | | Crystal structure of the N-terminal HTH DNA-binding domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
2R1J
 
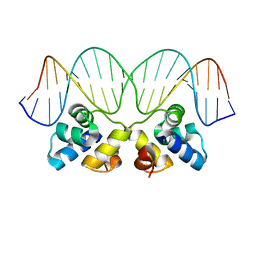 | | Crystal Structure of the P22 c2 Repressor protein in complex with the synthetic operator 9T | | Descriptor: | 5'-D(*DCP*DAP*DTP*DTP*DTP*DAP*DAP*DGP*DAP*DTP*DAP*DTP*DCP*DTP*DTP*DAP*DAP*DAP*DTP*DA)-3', 5'-D(*DTP*DAP*DTP*DTP*DTP*DAP*DAP*DGP*DAP*DTP*DAP*DTP*DCP*DTP*DTP*DAP*DAP*DAP*DTP*DG)-3', Repressor protein C2 | | Authors: | Williams, L.D, Koudelka, G.B, Watkins, D, Hsiao, C, Woods, K. | | Deposit date: | 2007-08-22 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | P22 c2 repressor-operator complex: mechanisms of direct and indirect readout
Biochemistry, 47, 2008
|
|
1LRP
 
 | |
1PER
 
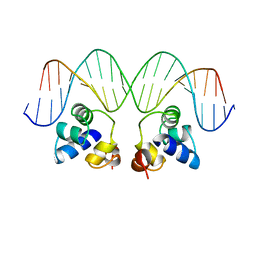 | |
1LMB
 
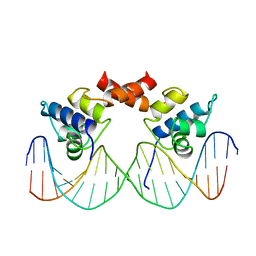 | | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Beamer, L.J, Pabo, C.O. | | Deposit date: | 1991-11-05 | | Release date: | 1991-11-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A crystal structure of the lambda repressor-operator complex.
J.Mol.Biol., 227, 1992
|
|
1PRA
 
 | |
1X57
 
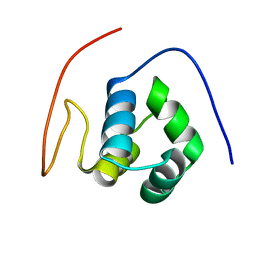 | | Solution structures of the HTH domain of human EDF-1 protein | | Descriptor: | Endothelial differentiation-related factor 1 | | Authors: | Nameki, N, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the HTH domain of human EDF-1 protein
To be Published
|
|
1SQ8
 
 | |
3CEC
 
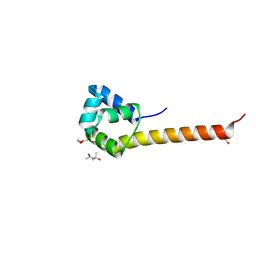 | |
4OB4
 
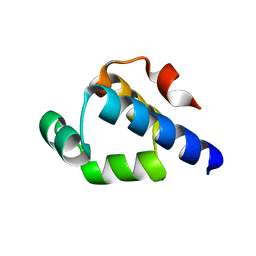 | | Structure of the S. venezulae BldD DNA-binding domain | | Descriptor: | Putative DNA-binding protein | | Authors: | schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
3CRO
 
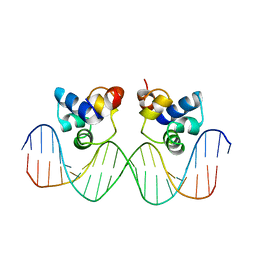 | | THE PHAGE 434 CRO/OR1 COMPLEX AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3'), PROTEIN (434 CRO) | | Authors: | Mondragon, A, Harrison, S.C. | | Deposit date: | 1990-07-06 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The phage 434 Cro/OR1 complex at 2.5 A resolution.
J.Mol.Biol., 219, 1991
|
|
3LFP
 
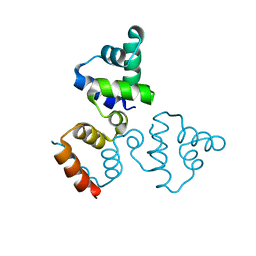 | |
3LIS
 
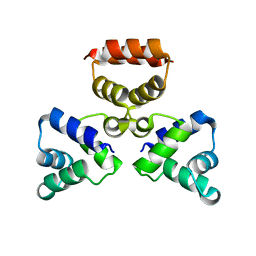 | |
4PU8
 
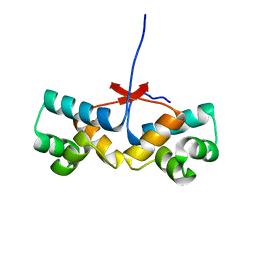 | | Shewanella oneidensis Toxin Antitoxin System Antitoxin Protein HipB Resolution 2.35 | | Descriptor: | Toxin-antitoxin system antidote transcriptional repressor Xre family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
1R69
 
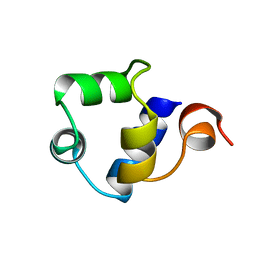 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
3CLC
 
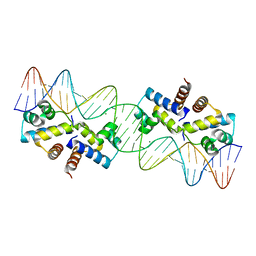 | | Crystal Structure of the Restriction-Modification Controller Protein C.Esp1396I Tetramer in Complex with its Natural 35 Base-Pair Operator | | Descriptor: | 35-MER, MAGNESIUM ION, Regulatory protein | | Authors: | McGeehan, J.E, Streeter, S.D, Thresh, S.J, Ball, N, Ravelli, R.B, Kneale, G.G. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the genetic switch that regulates the expression of restriction-modification genes.
Nucleic Acids Res., 36, 2008
|
|
