6NH5
 
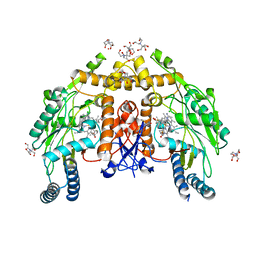 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 6-(3-fluoro-5-(2-((2R,4S)-4-fluoro-1-methylpyrrolidin-2-yl)ethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(3-fluoro-5-{2-[(2R,4S)-4-fluoro-1-methylpyrrolidin-2-yl]ethyl}phenyl)ethyl]-4-methylpyridin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
7V1C
 
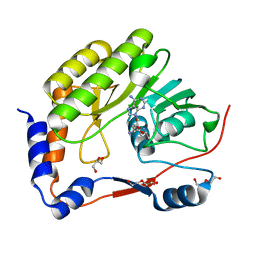 | |
7V1G
 
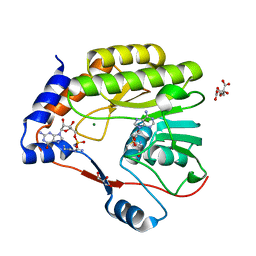 | |
8PQA
 
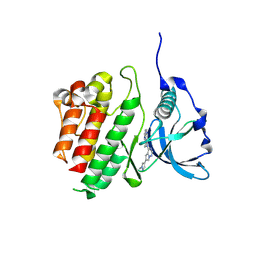 | | c-KIT kinase domain in complex with avapritinib derivative 4 | | Descriptor: | 6-(1-methylpyrazol-4-yl)-4-(4-pyrimidin-2-ylpiperazin-1-yl)pyrrolo[2,1-f][1,2,4]triazine, Mast/stem cell growth factor receptor Kit | | Authors: | Teuber, A, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-07-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Avapritinib-based SAR studies unveil a binding pocket in KIT and PDGFRA.
Nat Commun, 15, 2024
|
|
8B4J
 
 | | Rfa1-N-terminal domain in complex with phosphorylated Ddc2 | | Descriptor: | 1,2-ETHANEDIOL, DNA damage checkpoint protein LCD1, Replication factor A protein 1, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A DNA damage-induced phosphorylation circuit enhances Mec1 ATR Ddc2 ATRIP recruitment to Replication Protein A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7V1E
 
 | |
7V1F
 
 | |
8U8Q
 
 | | V290N/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5OD4
 
 | |
8S6H
 
 | |
7V45
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with para-bromotoluene. | | Descriptor: | 1-bromanyl-4-methyl-benzene, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
8U8R
 
 | | Y229F/V290N/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8S
 
 | | Y229F/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
9DCR
 
 | | Structure of the TelA-associated type VII secretion system chaperone SIR_0168 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DUF4176 domain-containing protein | | Authors: | Grebenc, D.W, Kim, Y, Whitney, J.C. | | Deposit date: | 2024-08-27 | | Release date: | 2025-11-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A widespread family of molecular chaperones promotes the intracellular stability of type VIIb secretion system-exported toxins.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6Q35
 
 | | Crystal structure of GES-5 beta-lactamase in complex with boronic inhibitor cpd 3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
4ZRM
 
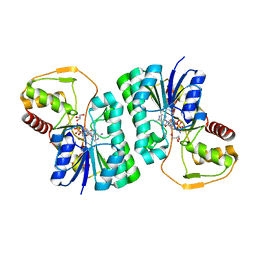 | |
6F3D
 
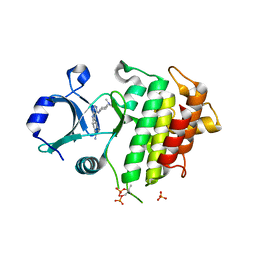 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 4-[4-[[4-(dimethylamino)cyclohexyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]cyclohexane-1-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
7S6M
 
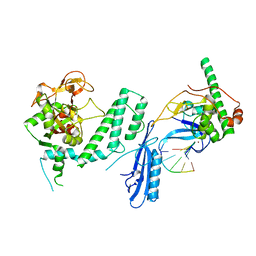 | |
8JH3
 
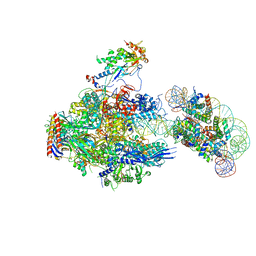 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
9KV0
 
 | | Cryo-EM structure of human G6PT in complex with chlorogenic acid | | Descriptor: | (1S,3R,4R,5R)-3-[(E)-3-[3,4-bis(oxidanyl)phenyl]prop-2-enoyl]oxy-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, Glucose-6-phosphate exchanger SLC37A4 | | Authors: | Jiang, D.H, Xia, Z.Y. | | Deposit date: | 2024-12-04 | | Release date: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for transport and inhibition of the human glucose-6-phosphate transporter G6PT.
Nat Commun, 16, 2025
|
|
6Q9H
 
 | |
8JH4
 
 | | RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
7PGJ
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-decarboxylate TamK and 10-hydroxylase RdmB, together with a single point mutation F297G | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-7-methoxy-2,4,5-tris(oxidanyl)-6,11-bis(oxidanylidene)-3,4-dihydro-1H-tetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7B1E
 
 | | BACE1 IN COMPLEX WITH compound 3 (NB-641) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(4~{S})-2-azanyl-4-methyl-5,6-dihydro-1,3-thiazin-4-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
6HM5
 
 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a RAD9 phosphopeptide | | Descriptor: | Cell cycle checkpoint control protein RAD9A, DNA topoisomerase II binding protein 1 | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.330038 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
