3WXJ
 
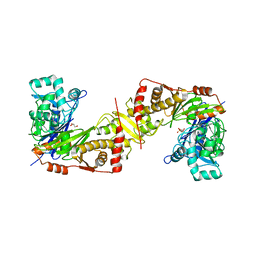 | | Crystal structure of trypanosoma brucei gambiense glycerol kinase in complex with glycerol 3-phosphate | | Descriptor: | GLYCEROL, Glycerol kinase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Kido, Y, Tsuge, C, Nara, T, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Kita, K, Harada, S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for the reverse reaction of African human trypanosomes glycerol kinase.
Mol.Microbiol., 94, 2014
|
|
3AEZ
 
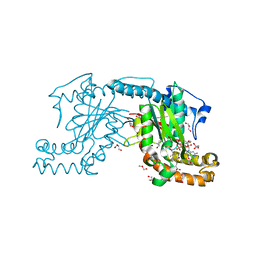 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with GDP and Phosphopantothenate | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanine, ... | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
3AF0
 
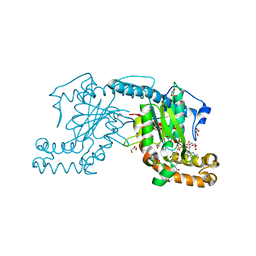 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with GDP and Pantothenate | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-19 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
3AF4
 
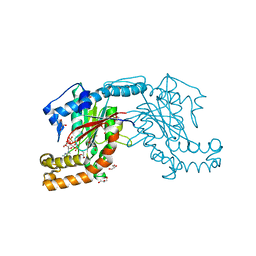 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with GMPPCP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Pantothenate kinase | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
3AF2
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with AMPPCP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Pantothenate kinase | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
3AF1
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with GDP | | Descriptor: | CHLORIDE ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-19 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
3AF3
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with GMPPCP and Pantothenate | | Descriptor: | GLYCEROL, PANTOTHENOIC ACID, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
2ZSE
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with AMPPCP and Pantothenate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, GLYCEROL, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZS9
 
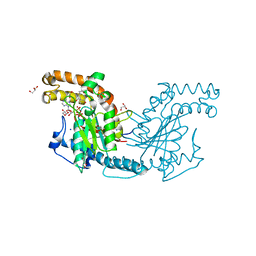 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with ADP and Pantothenate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-04 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4XR7
 
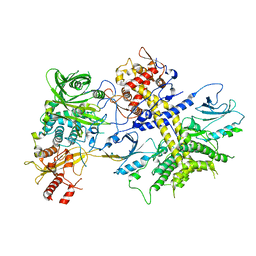 | | Structure of the Saccharomyces cerevisiae PAN2-PAN3 core complex | | Descriptor: | PAB-dependent poly(A)-specific ribonuclease subunit PAN2, PAB-dependent poly(A)-specific ribonuclease subunit PAN3 | | Authors: | Schafer, I.B, Rode, M, Bonneau, F, Schussler, S, Conti, E. | | Deposit date: | 2015-01-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.796 Å) | | Cite: | The structure of the Pan2-Pan3 core complex reveals cross-talk between deadenylase and pseudokinase.
Nat. Struct. Mol. Biol., 21, 2014
|
|
3W54
 
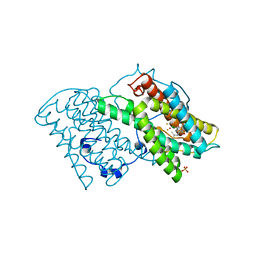 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with colletochlorin B | | Descriptor: | 3-chloro-5-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-4,6-dihydroxy-2-methylbenzaldehyde, Alternative oxidase, mitochondrial, ... | | Authors: | Shiba, T, Kido, Y, Sakamoto, K, Inaoka, D.K, Tsuge, C, Tatsumi, R, Takahashi, G, Balogun, E.O, Nara, T, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Saimoto, H, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the trypanosome cyanide-insensitive alternative oxidase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2ZSD
 
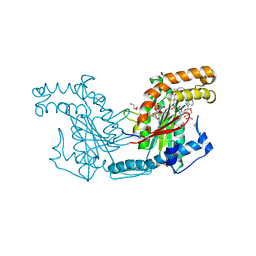 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with Coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZSA
 
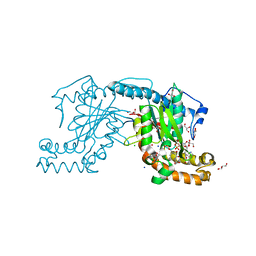 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with ADP and Phosphopantothenate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-04 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZSB
 
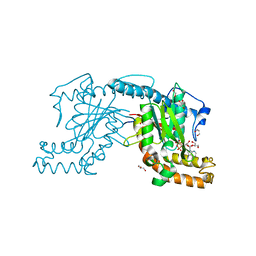 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with ADP, obtained through soaking of native enzyme crystals with the ligand | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Pantothenate kinase | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-04 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZS8
 
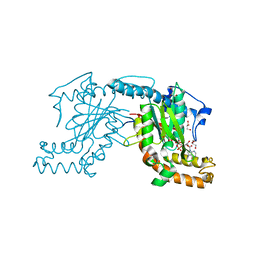 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) co-crystallized with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Pantothenate kinase | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZSF
 
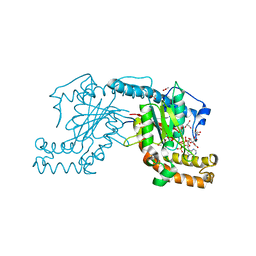 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with ATP and ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZS7
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with citrate anion | | Descriptor: | CITRATE ANION, GLYCEROL, Pantothenate kinase, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1MS1
 
 | | Monoclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
8RVR
 
 | |
4CJX
 
 | | The crystal structure of Trypanosoma brucei N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) complexed with NADP cofactor and inhibitor | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC, ... | | Authors: | Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2013-12-23 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of 2,4-Diamino-6-Oxo-1,6-Dihydropyrimidin-5-Yl Ureido Based Inhibitors of Trypanosoma Brucei Fold and Testing for Antiparasitic Activity.
J.Med.Chem., 58, 2015
|
|
7OQR
 
 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | ACETATE ION, Ascorbate peroxidase, GLYCEROL, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
8RTF
 
 | |
7FV7
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z1929967066 | | Descriptor: | (3R)-4-(furan-2-carbonyl)-3-methyl-N-(propan-2-yl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FV8
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z964297186 | | Descriptor: | 4-(3-chlorobenzoyl)-N-[3-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-3-oxopropyl]-1,4-diazepane-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FVN
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z371875396 | | Descriptor: | N-(propan-2-yl)-4-(thiophene-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
