1GAD
 
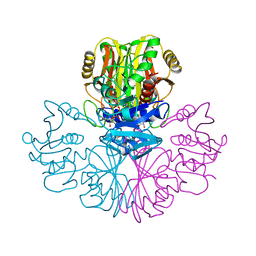 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
1GAE
 
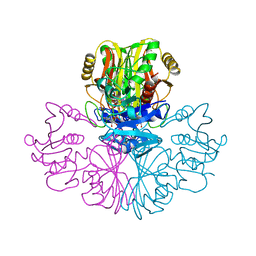 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
1SPE
 
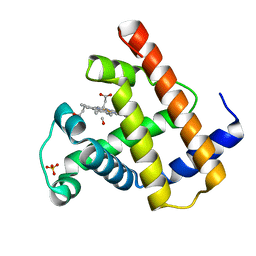 | | SPERM WHALE NATIVE CO MYOGLOBIN AT PH 4.0, TEMP 4C | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1RVV
 
 | | SYNTHASE/RIBOFLAVIN SYNTHASE COMPLEX OF BACILLUS SUBTILIS | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, PHOSPHATE ION, RIBOFLAVIN SYNTHASE | | Authors: | Ritsert, K, Huber, R, Turk, D, Ladenstein, R, Schmidt-Baese, K, Bacher, A. | | Deposit date: | 1995-10-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: crystal structure analysis of reconstituted, icosahedral beta-subunit capsids with bound substrate analogue inhibitor at 2.4 A resolution.
J.Mol.Biol., 253, 1995
|
|
1FSS
 
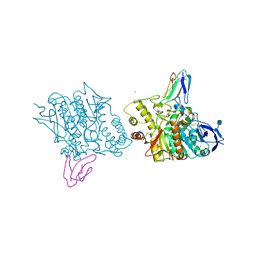 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH FASCICULIN-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, FASCICULIN II, ... | | Authors: | Harel, M, Kleywegt, G.J, Silman, I, Sussman, J.L. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an acetylcholinesterase-fasciculin complex: interaction of a three-fingered toxin from snake venom with its target.
Structure, 3, 1995
|
|
1MMC
 
 | | 1H NMR STUDY OF THE SOLUTION STRUCTURE OF AC-AMP2 | | Descriptor: | ANTIMICROBIAL PEPTIDE 2 | | Authors: | Martins, J.C, Maes, D, Loris, R, Pepermans, H.A.M, Wyns, L, Willem, R, Verheyden, P. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | H NMR study of the solution structure of Ac-AMP2, a sugar binding antimicrobial protein isolated from Amaranthus caudatus.
J.Mol.Biol., 258, 1996
|
|
1MIT
 
 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
1DDP
 
 | | Solution structure of a CISPLATIN-INDUCED [CATAGCTATG]2 Interstrand cross-link | | Descriptor: | Cisplatin, DNA (5'-D(*CP*AP*TP*AP*GP*CP*TP*AP*TP*G)-3') | | Authors: | Zhu, L, Huang, H, Reid, B.R, Drobny, G.P, Hopkins, P.B. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cisplatin-induced DNA interstrand cross-link.
Science, 270, 1995
|
|
1XRA
 
 | | CRYSTAL STRUCTURE OF S-ADENOSYLMETHIONINE SYNTHETASE | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
1XRB
 
 | | S-adenosylmethionine synthetase (MAT, ATP: L-methionine S-adenosyltransferase, E.C.2.5.1.6) in which MET residues are replaced with selenomethionine residues (MSE) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
1XRC
 
 | | CRYSTAL STRUCTURE OF S-ADENOSYLMETHIONINE SYNTHETASE | | Descriptor: | COBALT (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
1AAB
 
 | | NMR STRUCTURE OF RAT HMG1 HMGA FRAGMENT | | Descriptor: | HIGH MOBILITY GROUP PROTEIN | | Authors: | Hardman, C.H, Broadhurst, R.W, Raine, A.R.C, Grasser, K.D, Thomas, J.O, Laue, E.D. | | Deposit date: | 1995-10-28 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 34, 1995
|
|
2DLD
 
 | | D-LACTATE DEHYDROGENASE COMPLEXED WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Dunn, C.R, Holbrook, J.J. | | Deposit date: | 1995-10-28 | | Release date: | 1996-03-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dehydrogenases Engineering to Correct Substrate Inhibition in a Commercial Dehydrogenase
To be Published
|
|
1ORC
 
 | |
1UDH
 
 | |
1UDI
 
 | |
1UMS
 
 | | STROMELYSIN-1 CATALYTIC DOMAIN WITH HYDROPHOBIC INHIBITOR BOUND, PH 7.0, 32OC, 20 MM CACL2, 15% ACETONITRILE; NMR ENSEMBLE OF 20 STRUCTURES | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1UMT
 
 | | Stromelysin-1 catalytic domain with hydrophobic inhibitor bound, ph 7.0, 32oc, 20 mm cacl2, 15% acetonitrile; nmr average of 20 structures minimized with restraints | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1YRN
 
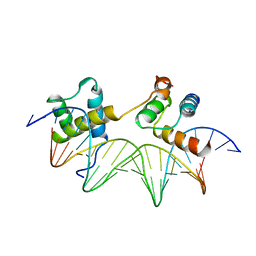 | | CRYSTAL STRUCTURE OF THE MATA1/MATALPHA2 HOMEODOMAIN HETERODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*TP*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*TP*AP*TP*TP*AP*C P*AP*TP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*AP*TP*GP*TP*AP*AP*TP*AP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT A1 HOMEODOMAIN), ... | | Authors: | Li, T, Stark, M.R, Johnson, A.D, Wolberger, C. | | Deposit date: | 1995-11-02 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA.
Science, 270, 1995
|
|
239D
 
 | |
238D
 
 | |
1AFD
 
 | |
1AGG
 
 | | THE SOLUTION STRUCTURE OF OMEGA-AGA-IVB, A P-TYPE CALCIUM CHANNEL ANTAGONIST FROM THE VENOM OF AGELENOPSIS APERTA | | Descriptor: | OMEGA-AGATOXIN-IVB | | Authors: | Reily, M.D, Thanabal, V, Adams, M.E. | | Deposit date: | 1995-11-03 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | The solution structure of omega-Aga-IVB, a P-type calcium channel antagonist from venom of the funnel web spider, Agelenopsis aperta.
J.Biomol.NMR, 5, 1995
|
|
1AFA
 
 | |
1XXA
 
 | | C-TERMINAL DOMAIN OF ESCHERICHIA COLI ARGININE REPRESSOR/ L-ARGININE COMPLEX; PB DERIVATIVE | | Descriptor: | ARGININE, ARGININE REPRESSOR, LEAD (II) ION | | Authors: | Van Duyne, G.D, Ghosh, G, Maas, W.K, Sigler, P.B. | | Deposit date: | 1995-11-03 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli.
J.Mol.Biol., 256, 1996
|
|
