8R02
 
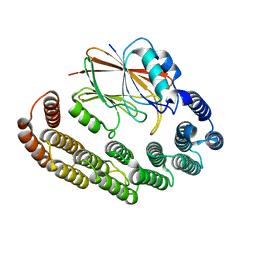 | | Crystal structure of the retromer complex VPS29/VPS35 with the ligand bis-1,3-phenyl guanylhydrazone, 2a | | Descriptor: | Bis-1,3-phenyl guanylhydrazon, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Milani, M, Fagnani, E. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stabilization of the retromer complex: Analysis of novel binding sites of bis-1,3-phenyl guanylhydrazone 2a to the VPS29/VPS35 interface.
Comput Struct Biotechnol J, 23, 2024
|
|
7BP5
 
 | |
7MEF
 
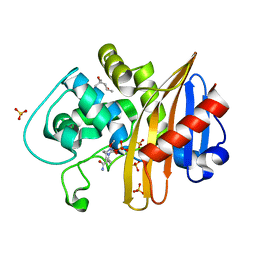 | | CDD-1 beta-lactamase in imidazole/MPD 10 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEE
 
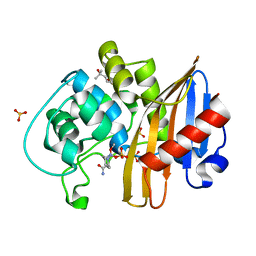 | | CDD-1 beta-lactamase in imidazole/MPD 6 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEC
 
 | | CDD-1 beta-lactamase in imidazole/MPD 4 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEG
 
 | | CDD-1 beta-lactamase in imidazole/MPD 30 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEB
 
 | | CDD-1 beta-lactamase in imidazole/MPD 2 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MED
 
 | | CDD-1 beta-lactamase in imidazole/MPD 5 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEH
 
 | | CDD-1 beta-lactamase in imidazole/MPD 60 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
9K6G
 
 | | Structure of human LINE-1 ORF2p with endogenous dsDNA and RNA/cDNA hybrid | | Descriptor: | Complementary DNA, Genomic DNA (bottom strand), Genomic DNA (top strand), ... | | Authors: | Jin, W, Yu, C, Xu, R.M. | | Deposit date: | 2024-10-22 | | Release date: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of DNA targeting by human LINE-1.
Science, 390, 2025
|
|
9K6H
 
 | | Structure of human LINE-1 ORF2p with endogenous DNA and RNA/cDNA hybrid bound to dNTP and Mn2+ | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Complementary DNA, Genomic DNA (bottom strand), ... | | Authors: | Jin, W, Yu, C, Xu, R.M. | | Deposit date: | 2024-10-22 | | Release date: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of DNA targeting by human LINE-1.
Science, 390, 2025
|
|
6YU1
 
 | | CLK3 bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
9K6I
 
 | | Structure of full-length human LINE-1 ORF2p with endogenous DNA and RNA/cDNA hybrid | | Descriptor: | Complementary DNA, Genomic DNA (bottom strand), Genomic DNA (top strand), ... | | Authors: | Jin, W, Yu, C, Xu, R.M. | | Deposit date: | 2024-10-22 | | Release date: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of DNA targeting by human LINE-1.
Science, 390, 2025
|
|
6YUY
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 471 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-5-(trifluoromethyl)-1~{H}-pyrrole-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6WRK
 
 | | Crystal structure of 3rd-generation Mj 3-nitro-tyrosine tRNA synthetase ("A7") bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
9GSS
 
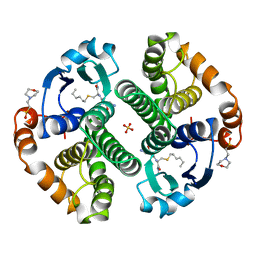 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH S-HEXYL GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, S-HEXYLGLUTATHIONE, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
6WPJ
 
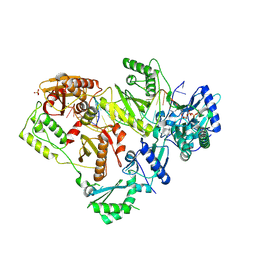 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and d4T | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA Primer 21-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Post-Catalytic Complexes with Emtricitabine or Stavudine and HIV-1 Reverse Transcriptase Reveal New Mechanistic Insights for Nucleotide Incorporation and Drug Resistance.
Molecules, 25, 2020
|
|
7UN0
 
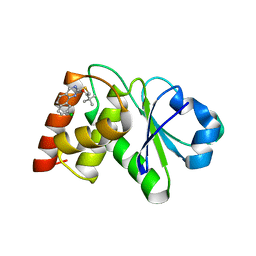 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-chloro-5,6-dihydrobenzo[h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 1-[(9-chlorobenzo[h]quinazolin-2-yl)sulfanyl]-3,3-dimethylbutan-2-one, Dual specificity protein phosphatase 10 | | Authors: | Gannam, Z.T.K, Jamali, H, Lolis, E, Anderson, K.S, Ellman, J.A, Bennett, A.M. | | Deposit date: | 2022-04-08 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Defining the structure-activity relationship for a novel class of allosteric MKP5 inhibitors.
Eur.J.Med.Chem., 243, 2022
|
|
7UMU
 
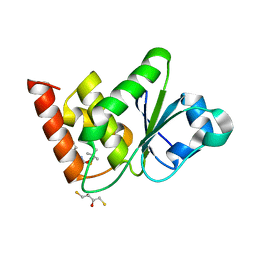 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((5,6-dihydrobenzo[h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 1-[(benzo[h]quinazolin-2-yl)sulfanyl]-3,3-dimethylbutan-2-one, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dual specificity protein phosphatase 10 | | Authors: | Gannam, Z.T.K, Jamali, H, Lolis, E, Anderson, K.S, Ellman, J.A, Bennett, A.M. | | Deposit date: | 2022-04-07 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Defining the structure-activity relationship for a novel class of allosteric MKP5 inhibitors.
Eur.J.Med.Chem., 243, 2022
|
|
6O4D
 
 | | Structure of ALDH7A1 mutant W175A complexed with L-pipecolic acid | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
7BUH
 
 | | Reduced ferredoxin of carbazole 1,9a-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin CarAc, ... | | Authors: | Matsuzawa, J, Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2020-04-06 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Reduced ferredoxin of carbazole 1,9a-dioxygenase
To Be Published
|
|
7BUI
 
 | | Complex of reduced oxygenase and oxidized ferredoxin in carbazole 1,9a- dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Matsuzawa, J, Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2020-04-06 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Complex of reduced oxygenase and oxidized ferredoxin in carbazole 1,9a- dioxygenase
To Be Published
|
|
6HIP
 
 | | Structure of SPF45 UHM bound to HIV-1 Rev ULM | | Descriptor: | HIV-1 Rev (41-49), SODIUM ION, Splicing factor 45, ... | | Authors: | Pabis, M, Corsini, L, Sattler, M. | | Deposit date: | 2018-08-30 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Modulation of HIV-1 gene expression by binding of a ULM motif in the Rev protein to UHM-containing splicing factors.
Nucleic Acids Res., 47, 2019
|
|
5MZ3
 
 | | P38 ALPHA MUTANT C162S IN COMPLEX WITH CMPD2 [N-(4-Methyl-3-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)-3-(trifluoromethyl)benzamide] | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Cowan-Jacob, S.W, Scheufler, C. | | Deposit date: | 2017-01-30 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6P3B
 
 | |
