1DCU
 
 | | REDOX SIGNALING IN THE CHLOROPLAST: STRUCTURE OF OXIDIZED PEA FRUCTOSE-1,6-BISPHOSPHATE PHOSPHATASE | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Chiadmi, M, Navaza, A, Miginiac-Maslow, M, Jacquot, J.P, Cherfils, J. | | Deposit date: | 1999-11-05 | | Release date: | 1999-12-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase.
EMBO J., 18, 1999
|
|
1DCV
 
 | |
1DCW
 
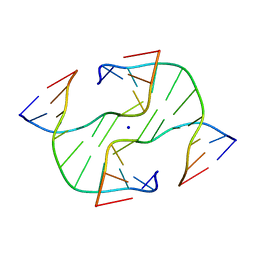 | | STRUCTURE OF A FOUR-WAY JUNCTION IN AN INVERTED REPEAT SEQUENCE. | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3'), SODIUM ION | | Authors: | Eichman, B.F, Vargason, J.M, Mooers, B.H.M, Ho, P.S. | | Deposit date: | 1999-11-05 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DCY
 
 | |
1DCZ
 
 | |
1DD1
 
 | |
1DD2
 
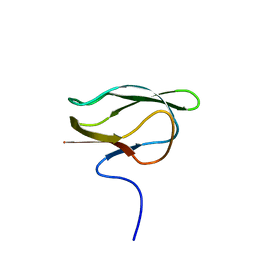 | |
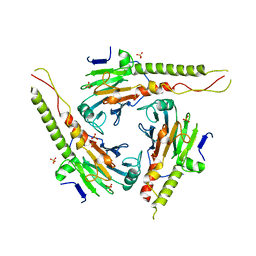
1DD3
 
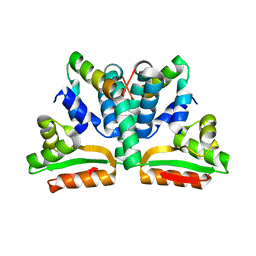 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L12 FROM THERMOTOGA MARITIMA | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12 | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
EMBO J., 19, 2000
|
|
1DD4
 
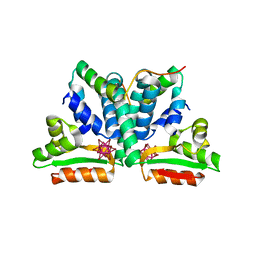 | | Crystal structure of ribosomal protein l12 from thermotoga maritim | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12, HEXATANTALUM DODECABROMIDE | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
Embo J., 19, 2000
|
|
1DD5
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA RIBOSOME RECYCLING FACTOR, RRF | | Descriptor: | ACETIC ACID, RIBOSOME RECYCLING FACTOR | | Authors: | Selmer, M, Al-Karadaghi, S, Hirokawa, G, Kaji, A, Liljas, A. | | Deposit date: | 1999-11-08 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic.
Science, 286, 1999
|
|
1DD6
 
 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | Descriptor: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DD7
 
 | |
1DD8
 
 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I FROM ESCHERICHIA COLI | | Descriptor: | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I | | Authors: | Olsen, J.G, Kadziola, A, von Wettstein-Knowles, P, Siggaard-Andersen, M, Lindquist, Y, Larsen, S. | | Deposit date: | 1999-11-09 | | Release date: | 1999-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structure of beta-ketoacyl [acyl carrier protein] synthase I.
FEBS Lett., 460, 1999
|
|
1DD9
 
 | | STRUCTURE OF THE DNAG CATALYTIC CORE | | Descriptor: | DNA PRIMASE, STRONTIUM ION | | Authors: | Keck, J.L, Roche, D.D, Lynch, A.S, Berger, J.M. | | Deposit date: | 1999-11-09 | | Release date: | 2000-04-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA polymerase domain of E. coli primase.
Science, 287, 2000
|
|
1DDB
 
 | | STRUCTURE OF MOUSE BID, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (BID) | | Authors: | Mcdonnell, J.M, Fushman, D, Milliman, C, Korsmeyer, S.J, Cowburn, D. | | Deposit date: | 1999-02-19 | | Release date: | 1999-08-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists.
Cell(Cambridge,Mass.), 96, 1999
|
|
1DDE
 
 | | STRUCTURE OF THE DNAG CATALYTIC CORE | | Descriptor: | DNA PRIMASE, YTTRIUM ION | | Authors: | Keck, J.L, Roche, D.D, Lynch, A.S, Berger, J.M. | | Deposit date: | 1999-11-09 | | Release date: | 2000-04-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RNA polymerase domain of E. coli primase.
Science, 287, 2000
|
|
1DDF
 
 | | FAS DEATH DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FAS | | Authors: | Huang, B, Eberstadt, M, Olejniczak, E, Meadows, R.P, Fesik, S. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain.
Nature, 384, 1996
|
|
1DDG
 
 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, SULFITE REDUCTASE (NADPH) FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1DDH
 
 | |
1DDI
 
 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFITE REDUCTASE [NADPH] FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1DDJ
 
 | | CRYSTAL STRUCTURE OF HUMAN PLASMINOGEN CATALYTIC DOMAIN | | Descriptor: | PLASMINOGEN | | Authors: | Wang, X, Terzyan, S, Tang, J, Loy, J, Lin, X, Zhang, X. | | Deposit date: | 1999-11-10 | | Release date: | 2000-02-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human plasminogen catalytic domain undergoes an unusual conformational change upon activation.
J.Mol.Biol., 295, 2000
|
|
1DDK
 
 | | CRYSTAL STRUCTURE OF IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, ZINC ION | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DDL
 
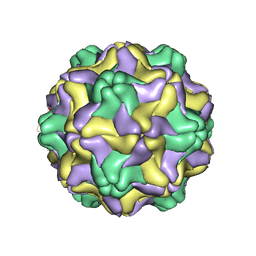 | | DESMODIUM YELLOW MOTTLE TYMOVIRUS | | Descriptor: | DESMODIUM YELLOW MOTTLE VIRUS, RNA (5'-R(P*UP*U)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Larson, S.B, Day, J, Canady, M.A, Greenwood, A, McPherson, A. | | Deposit date: | 1999-11-10 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Refined structure of desmodium yellow mottle tymovirus at 2.7 A resolution.
J.Mol.Biol., 301, 2000
|
|
1DDM
 
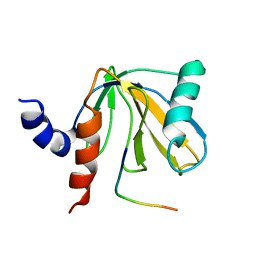 | | SOLUTION STRUCTURE OF THE NUMB PTB DOMAIN COMPLEXED TO A NAK PEPTIDE | | Descriptor: | NUMB ASSOCIATE KINASE, NUMB PROTEIN | | Authors: | Zwahlen, C, Li, S.C, Kay, L.E, Pawson, T, Forman-Kay, J.D. | | Deposit date: | 1999-11-11 | | Release date: | 2000-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Multiple modes of peptide recognition by the PTB domain of the cell fate determinant Numb.
EMBO J., 19, 2000
|
|
1DDN
 
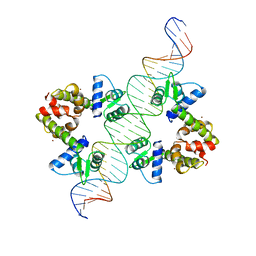 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT)/TOX DNA OPERATOR COMPLEX | | Descriptor: | 33 BASE DNA CONTAINING TOXIN OPERATOR, DIPHTHERIA TOX REPRESSOR, NICKEL (II) ION | | Authors: | White, A, Ding, X, Vanderspek, J.C, Murphy, J.R, Ringe, D. | | Deposit date: | 1998-06-23 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex.
Nature, 394, 1998
|
|
