8HAW
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8HMU
 
 | | Crystal Structure of PKM2 mutant R516C | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
5YZC
 
 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor compound (AS-48) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitro-2-[(phenylacetyl)amino]benzamide, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8HDU
 
 | |
8HDV
 
 | |
5YZM
 
 | | Crystal structure of S9 peptidase (inactive form) from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Acyl-peptide hydrolase, putative | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
5ZC4
 
 | |
8H99
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
8H9C
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 4 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
5Z37
 
 | |
5ZD4
 
 | | Crystal structure of MBP-fused BIL1/BZR1 in complex with double-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Miyakawa, T, Xu, Y, Nakamura, A, Hirabayashi, K, Tanokura, M. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for brassinosteroid response by BIL1/BZR1.
Nat Plants, 4, 2018
|
|
5ZIS
 
 | | Crystal structure of Mn-ProtoporphyrinIX-reconstituted P450BM3 | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, MANGANESE PROTOPORPHYRIN IX | | Authors: | Omura, K, Aiba, Y, Onoda, H, Sugimoto, H, Shoji, O, Watanabe, Y. | | Deposit date: | 2018-03-17 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Chem. Commun. (Camb.), 54, 2018
|
|
8H98
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 1 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
5ZM3
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AndA with preandiloid B | | Descriptor: | (6aS,8aR,12aS,12bR,13aR)-5,6a,9,9,12a,13a-hexamethyl-7,8,8a,9,11,12,12a,12b,13,13a-decahydro-3H-benzo[a]furo[3,4-j]xanthene-3,4,10(1H,6aH)-trione, 2-OXOGLUTARIC ACID, Dioxygenase andA, ... | | Authors: | Nakashima, Y, Senda, T. | | Deposit date: | 2018-04-01 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and Computational Bases for Dramatic Skeletal Rearrangement in Anditomin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
8HAV
 
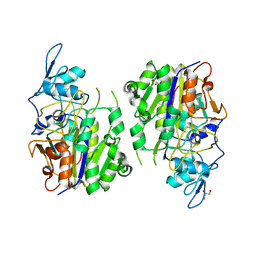 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-specific phospholipase C4 | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
5ZNH
 
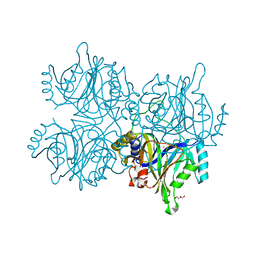 | | Catechol 2,3-dioxygenase with 4-methyl catechol from Diaphorobacter sp DS2 | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLCATECHOL, CALCIUM ION, ... | | Authors: | Mishra, K, Arya, C.K, Subramaniyan, R, Ramanathan, G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | catechol 2,3-dioxygenase with 4-methyl catechol from Diaphorobacter sp DS2
To Be Published
|
|
8H9B
 
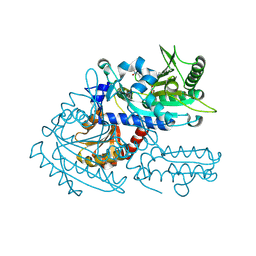 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 3 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
5ZJB
 
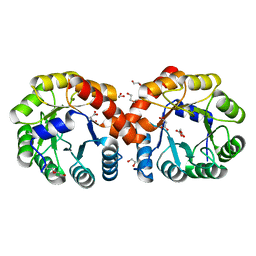 | |
5ZJQ
 
 | | Structure of AbdB/Exd complex bound to a 'Red14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*TP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Baburajendran, N, Zeiske, T, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
5ZKN
 
 | |
8H9A
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 2 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
8GYH
 
 | | Crystal structure of Fic25 (apo form) from Streptomyces ficellus | | Descriptor: | DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, IMIDAZOLE | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
5Z8Z
 
 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 6-[(3S)-3-azanylpiperidin-1-yl]sulfonyl-1-ethyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
5ZLV
 
 | |
