7OY8
 
 | | Cryo-EM structure of the Rhodospirillum rubrum RC-LH1 complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 2-azanyl-5-[(2~{E},6~{E},8~{E},10~{E},12~{E},14~{E},18~{E},22~{E},26~{E},30~{E},34~{E})-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,8,10,12,14,18,22,26,30,34,38-dodecaenyl]-3-methoxy-6-methyl-cyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Qian, P, Croll, T.I, Castro, H.P, Moriarty, N.W, sader, K, Hunter, C.N. | | Deposit date: | 2021-06-23 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the Rhodospirillum rubrum RC-LH1 complex at 2.5 angstrom.
Biochem.J., 478, 2021
|
|
3V3Y
 
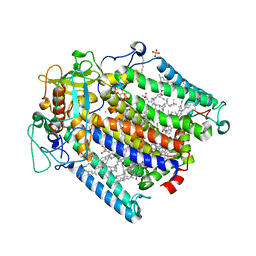 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Gabdulkhakov, A.G, Fufina, T.Y, Vasilieva, L.G, Shuvalov, V.A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The site-directed mutation I(L177)H in Rhodobacter sphaeroides reaction center affects coordination of P(A) and B(B) bacteriochlorophylls.
Biochim.Biophys.Acta, 1817, 2012
|
|
7MH4
 
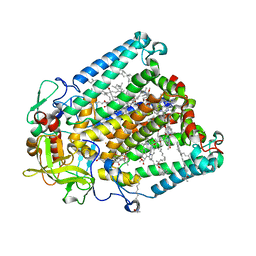 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-bromotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH9
 
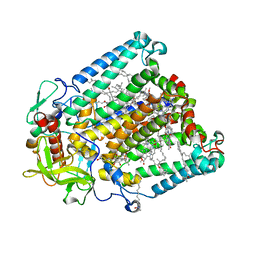 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-nitrotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH5
 
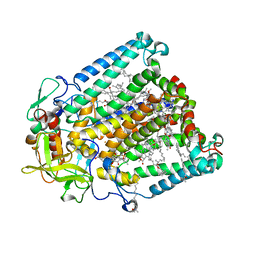 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-iodotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH8
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-methyltyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH3
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-chlorotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J.B, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MHA
 
 | |
4CAS
 
 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
2MQH
 
 | |
2WX5
 
 | | Hexa-coordination of a bacteriochlorophyll cofactor in the Rhodobacter sphaeroides reaction centre | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Marsh, M, Frolov, D, Crouch, L.I, Fyfe, P.K, Robert, B, van Grondelle, R, Jones, M.R, Hadfield, A.T. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Spectroscopic Consequences of Hexa-Coordination of a Bacteriochlorophyll Cofactor in the Rhodobacter Sphaeroides Reaction Centre
Biochemistry, 49, 2010
|
|
8VTM
 
 | |
8VTJ
 
 | |
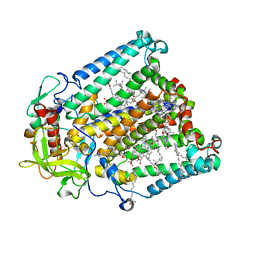
8VTK
 
 | |
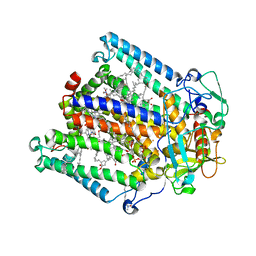
8VTO
 
 | |
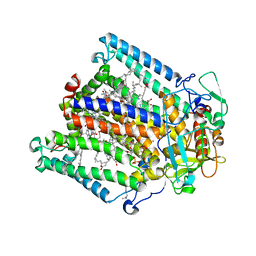
8VTL
 
 | |
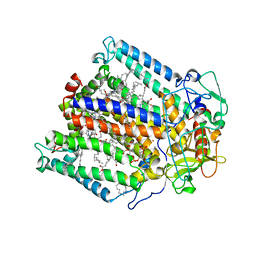
8VTN
 
 | |
7VRJ
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF Allochromatium tepidum | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Tani, K, Kobayashi, K, Hosogi, N, Ji, X.-C, Nagashima, S, Nagashima, K.V.P, Tsukatani, Y, Kanno, R, Hall, M, Yu, L.-J, Ishikawa, I, Okura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-10-23 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | A Ca 2+ -binding motif underlies the unusual properties of certain photosynthetic bacterial core light-harvesting complexes.
J.Biol.Chem., 298, 2022
|
|
2ZT9
 
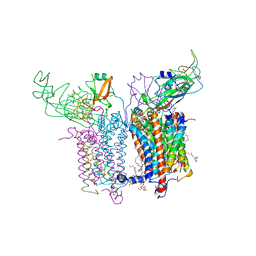 | | Crystal Structure of the Cytochrome b6f Complex from Nostoc sp. PCC 7120 | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, Apocytochrome f, ... | | Authors: | Craner, W.A, Baniulis, D, Yamashita, E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Function, Stability, and Chemical Modification of the Cyanobacterial Cytochrome b6f Complex from Nostoc sp. PCC 7120
J.Biol.Chem., 284, 2009
|
|
1EYS
 
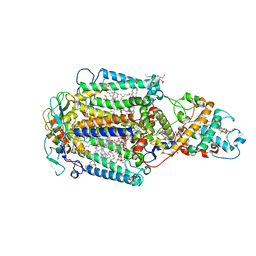 | | CRYSTAL STRUCTURE OF PHOTOSYNTHETIC REACTION CENTER FROM A THERMOPHILIC BACTERIUM, THERMOCHROMATIUM TEPIDUM | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Nogi, T, Fathir, I, Kobayashi, M, Nozawa, T, Miki, K. | | Deposit date: | 2000-05-08 | | Release date: | 2000-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of photosynthetic reaction center and high-potential iron-sulfur protein from Thermochromatium tepidum: thermostability and electron transfer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4AC5
 
 | | Lipidic sponge phase crystal structure of the Bl. viridis reaction centre solved using serial femtosecond crystallography | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Johansson, L.C, Arnlund, D, White, T.A, Katona, G, DePonte, D.P, Weierstall, U, Doak, R.B, Shoeman, R.L, Lomb, L, Malmerberg, E, Davidsson, J, Nass, K, Liang, M, Andreasson, J, Aquila, A, Bajt, S, Barthelmess, M, Barty, A, Bogan, M.J, Bostedt, C, Bozek, J.D, Caleman, C, Coffee, R, Coppola, N, Ekeberg, T, Epp, S.W, Erk, B, Fleckenstein, H, Foucar, L, Graafsma, H, Gumprecht, L, Hajdu, J, Hampton, C.Y, Hartmann, R, Hartmann, A, Hauser, G, Hirsemann, H, Holl, P, Hunter, M.S, Kassemeyer, S, Kimmel, N, Kirian, R.A, Maia, F.R.N.C, Marchesini, S, Martin, A.V, Reich, C, Rolles, D, Rudek, B, Rudenko, A, Schlichting, I, Schulz, J, Seibert, M.M, Sierra, R, Soltau, H, Starodub, D, Stellato, F, Stern, S, Struder, L, Timneanu, N, Ullrich, J, Wahlgren, W.Y, Wang, X, Weidenspointner, G, Wunderer, C, Fromme, P, Chapman, H.N, Spence, J.C.H, Neutze, R. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (8.2 Å) | | Cite: | Lipidic Phase Membrane Protein Serial Femtosecond Crystallography.
Nat.Methods, 9, 2012
|
|
7YML
 
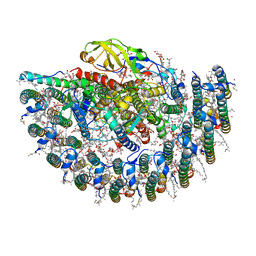 | | Structure of photosynthetic LH1-RC super-complex of Rhodobacter capsulatus | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tani, K, Kanno, R, Ji, X.-C, Satoh, I, Kobayashi, Y, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Mizoguchi, A, Humbel, B.M, Madigan, M.T, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Rhodobacter capsulatus forms a compact crescent-shaped LH1-RC photocomplex.
Nat Commun, 14, 2023
|
|
6AUC
 
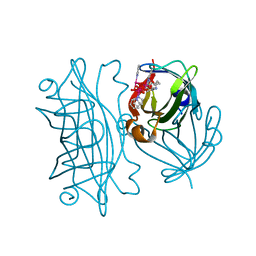 | | Artificial metalloproteins containing a Co4O4 active site - 2xm-Sav | | Descriptor: | N-biotin-C-Co4(mu3-O)4(Py)4(H2O)4-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurackal, J, Huerta-Lavorie, R, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
2Q5B
 
 | | High resolution structure of Plastocyanin from Phormidium laminosum | | Descriptor: | ACETATE ION, COPPER (II) ION, POTASSIUM ION, ... | | Authors: | Fromme, R, Bukhman-DeRuyter, Y.S, Grotjohann, I, Mi, H, Fromme, P. | | Deposit date: | 2007-05-31 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Plastocyanin at high resolution from Phormidium laminosum and the double mutant D44A, D45A
To be Published
|
|
2BZC
 
 | |
