3SBL
 
 | | Crystal Structure of New Delhi Metal-beta-lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CITRIC ACID | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3DAM
 
 | | Crystal Structure of Allene oxide synthase | | Descriptor: | Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3ZHW
 
 | | X-ray Crystallographic Structural Characteristics of Arabidopsis Hemoglobin I and their Functional Implications | | Descriptor: | NON-SYMBIOTIC HEMOGLOBIN 1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Mukhi, N, Dhindwal, S, Uppal, S, Kumar, P, Kaur, J, Kundu, S. | | Deposit date: | 2012-12-30 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | X-Ray Crystallographic Structural Characteristics of Arabidopsis Hemoglobin I and Their Functional Implications
Biochim.Biophys.Acta, 1834, 2013
|
|
3DKF
 
 | | Structure of MET receptor tyrosine kinase in complex with inhibitor SGX-523 | | Descriptor: | 6-{[6-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}quinoline, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Hendle, J. | | Deposit date: | 2008-06-24 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SGX523 is an exquisitely selective, ATP-competitive inhibitor of the MET receptor tyrosine kinase with antitumor activity in vivo.
Mol.Cancer Ther., 8, 2009
|
|
3DD8
 
 | | Carbonic anhydrase inhibitors. Interaction of the antitumor sulfamate EMD-486019 with twelve mammalian isoforms: kinetic and X-Ray crystallographic studies | | Descriptor: | 2-(cycloheptylmethyl)-1,1-dioxido-1-benzothiophen-6-yl sulfamate, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Temperini, C, Innocenti, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2008-06-05 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase inhibitors. Interaction of the antitumor sulfamate EMD 486019 with twelve mammalian carbonic anhydrase isoforms: Kinetic and X-ray crystallographic studies
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3SEQ
 
 | |
3DDS
 
 | |
2PHM
 
 | | STRUCTURE OF PHENYLALANINE HYDROXYLASE DEPHOSPHORYLATED | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE-4-HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
5U2T
 
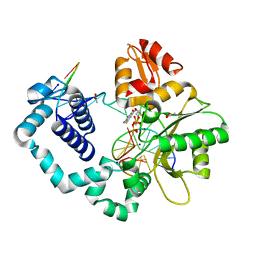 | |
3SIK
 
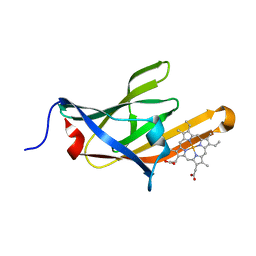 | |
4EGV
 
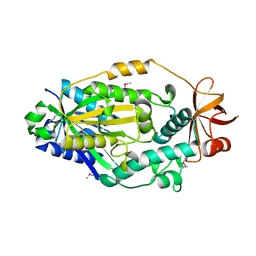 | |
5U3R
 
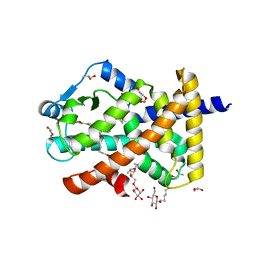 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 2 | | Descriptor: | 6-[2-({[4-(furan-2-yl)benzene-1-carbonyl](propan-2-yl)amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U43
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 12 | | Descriptor: | 6-(2-{[cyclopropyl(4'-methoxy[1,1'-biphenyl]-4-carbonyl)amino]methyl}phenoxy)hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3DPE
 
 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | CALCIUM ION, N-{[2-(2-amino-3,4-dioxocyclobut-1-en-1-yl)-1,2,3,4-tetrahydroisoquinolin-7-yl]methyl}-4-oxo-3,5,6,8-tetrahydro-4H-thiopyrano[4',3':4,5]thieno[2,3-d]pyrimidine-2-carboxamide 7,7-dioxide, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
4EDM
 
 | | Crystal structure of beta-parvin CH2 domain | | Descriptor: | 1,2-ETHANEDIOL, Beta-parvin | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
2PUT
 
 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | ACETATE ION, FRUCTOSE -6-PHOSPHATE, SODIUM ION, ... | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
5U6Y
 
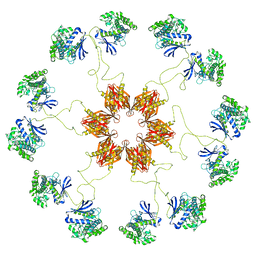 | | Pseudo-atomic model of the CaMKIIa holoenzyme. | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Myers, J, Reichow, S.L. | | Deposit date: | 2016-12-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | The CaMKII holoenzyme structure in activation-competent conformations.
Nat Commun, 8, 2017
|
|
1QGF
 
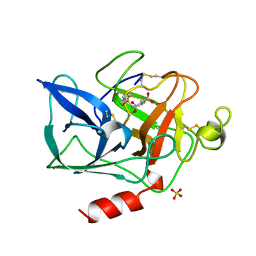 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3R, 4S)N-PARA-TOLUENESULPHONYL-3-ETHYL-4-(CARBOXYLIC ACID)PYRROLIDIN-2-ONE | | Descriptor: | (2S,3S)-3-FORMYL-2-({[(4-METHYLPHENYL)SULFONYL]AMINO}METHYL)PENTANOIC ACID, CALCIUM ION, ELASTASE, ... | | Authors: | Wilmouth, R.C, Kassamally, S, Westwood, N.J, Sheppard, R.J, Claridge, T.D.W, Wright, P.A, Pritchard, G.J, Schofield, C.J. | | Deposit date: | 1999-04-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic insights into the inhibition of serine proteases by monocyclic lactams.
Biochemistry, 38, 1999
|
|
3ZIT
 
 | |
3U9Z
 
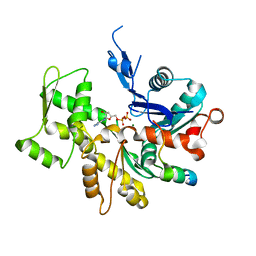 | | Crystal structure between actin and a protein construct containing the first beta-thymosin domain of drosophila ciboulot (residues 2-58) with the three mutations N26D/Q27K/D28S | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
3DRG
 
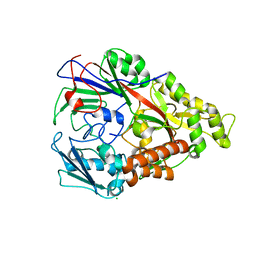 | | Lactococcal OppA complexed with bradykinin in the closed conformation | | Descriptor: | Bradykinin, CHLORIDE ION, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
5TQJ
 
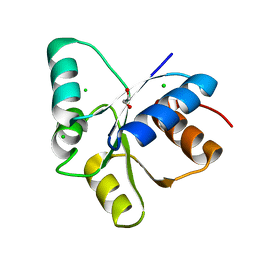 | |
3UAC
 
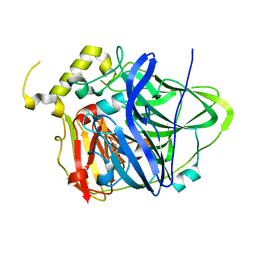 | | Multicopper Oxidase CueO mutant C500SE506Q (data4) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2PWN
 
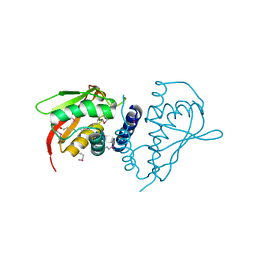 | |
4EKR
 
 | | T4 Lysozyme L99A/M102H with 2-Cyanophenol Bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 2-hydroxybenzonitrile, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering a model protein cavity to catalyze the Kemp elimination.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
