6ESP
 
 | |
6EWP
 
 | | Mus musculus CEP120 third C2 domain (C2C) | | Descriptor: | Centrosomal protein of 120 kDa, HEXAETHYLENE GLYCOL | | Authors: | van Breugel, M, al-Jassar, C. | | Deposit date: | 2017-11-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Disease-Associated Mutations in CEP120 Destabilize the Protein and Impair Ciliogenesis.
Cell Rep, 23, 2018
|
|
6EYA
 
 | |
8GKV
 
 | | Crystal structure of anti-adaptor IraP that regulates RpoS proteolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Shaw, G.X, Gan, J, Suburaman, P, Battesti, A, Zhou, Y.N, Wickner, S, Gottesman, S, Ji, X. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural and functional study of anti-adaptor IraP-mediated regulation of RpoS proteolysis
to be published
|
|
6EWH
 
 | |
6EYB
 
 | |
6F1N
 
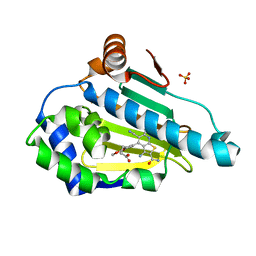 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 4-[5-[2-aminocarbonyl-3,6-bis(azanyl)-5-cyano-thieno[2,3-b]pyridin-4-yl]-2-methoxy-phenoxy]butanoic acid, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-11-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6F3O
 
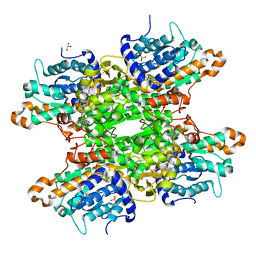 | |
6F6G
 
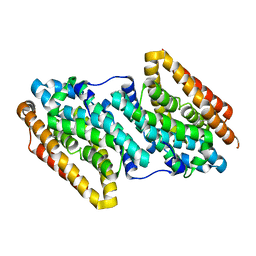 | |
6FFS
 
 | | Structure-based design and synthesis of macrocyclic human rhinovirus 3C protease inhibitors | | Descriptor: | 3C Protease, SULFATE ION, ~{N}-[(2~{S},5~{S},14~{S})-2-[(4-fluorophenyl)methyl]-5-(hydroxymethyl)-9-methyl-3,8,15-tris(oxidanylidene)-1,4,9-triazacyclopentadec-14-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wiesmann, C, Farady, C. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design and synthesis of macrocyclic human rhinovirus 3C protease inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6FCA
 
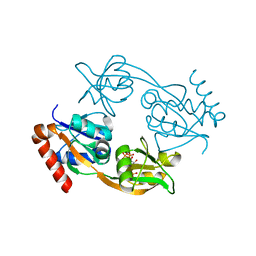 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ATP phosphoribosyltransferase | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
8GKF
 
 | |
6FC3
 
 | |
6FCC
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) | | Descriptor: | ATP phosphoribosyltransferase, L(+)-TARTARIC ACID | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FCW
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRATP | | Descriptor: | ATP phosphoribosyltransferase, MAGNESIUM ION, PHOSPHORIBOSYL ATP | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6F3T
 
 | | Crystal structure of the human TAF5-TAF6-TAF9 complex | | Descriptor: | CHLORIDE ION, Transcription initiation factor TFIID subunit 5, Transcription initiation factor TFIID subunit 6, ... | | Authors: | Haffke, M, Berger, I. | | Deposit date: | 2017-11-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chaperonin CCT checkpoint function in basal transcription factor TFIID assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6FDE
 
 | | Crystal Structure of the HHD2 Domain of Whirlin : 3-helix conformation | | Descriptor: | Whirlin | | Authors: | Delhommel, F, Cordier, F, Saul, F, Haouz, A, Wolff, N. | | Deposit date: | 2017-12-22 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural plasticity of the HHD2 domain of whirlin.
FEBS J., 285, 2018
|
|
6F6B
 
 | |
6FCJ
 
 | |
6FCT
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRPP and ATP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FJ4
 
 | |
6EZD
 
 | |
6F1K
 
 | | Structure of ARTD2/PARP2 WGR domain bound to double strand DNA without 5'phosphate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*CP*TP*AP*GP*CP*TP*AP*CP*GP*TP*AP*GP*CP*TP*AP*GP*GP*C)-3'), GLYCEROL, ... | | Authors: | Obaji, E, Haikarainen, T, Lehtio, L. | | Deposit date: | 2017-11-22 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for DNA break recognition by ARTD2/PARP2.
Nucleic Acids Res., 46, 2018
|
|
6F3N
 
 | |
8HDD
 
 | | Complex structure of catalytic, small, and a partial electron transfer subunits from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Sode, K. | | Deposit date: | 2022-11-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microgravity environment grown crystal structure information based engineering of direct electron transfer type glucose dehydrogenase.
Commun Biol, 5, 2022
|
|
