8X9G
 
 | | Crystal structure of CO dehydrogenase mutant in complex with BV | | Descriptor: | 1-(phenylmethyl)-4-[1-(phenylmethyl)pyridin-1-ium-4-yl]pyridin-1-ium, Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9F
 
 | | Crystal structure of CO dehydrogenase mutant in complex with EV | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-4-(1-ethylpyridin-1-ium-4-yl)pyridin-1-ium, Carbon monoxide dehydrogenase 2, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
5LHG
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-methyl-N-[4-[[4-(1-methylpiperidin-4-yl)oxyphenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-methyl-N-[4-[[4-[(1-methyl-4-piperidyl)oxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
6HAV
 
 | | Crystal structure of [Fe]-hydrogenase (Hmd) from Methanococcus aeolicus in complex with FeGP and methenyl-tetrahydromethanopterin (close form A) at 1.06 A resolution | | Descriptor: | 1-{4-[(6S,6aR,7R)-3-amino-6,7-dimethyl-1-oxo-1,2,5,6,6a,7-hexahydro-8H-imidazo[1,5-f]pteridin-10-ium-8-yl]phenyl}-1-deoxy-5-O-{5-O-[(S)-{[(1S)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, ... | | Authors: | Huang, G, Wagner, T, Wodrich, M.D, Ataka, K, Bill, E, Ermler, U, Hu, X, Shima, S. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The atomic-resolution crystal structure of activated [Fe]-hydrogenase
Nat Catal, 2019
|
|
6F4P
 
 | | Human JMJD5 in complex with MN, NOG and RPS6 (129-144) (complex-1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 40S ribosomal protein S6, JmjC domain-containing protein 5, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
6AO6
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
4EVM
 
 | | 1.5 Angstrom crystal structure of soluble domain of membrane-anchored thioredoxin family protein from Streptococcus pneumoniae strain Canada MDR_19A | | Descriptor: | Thioredoxin family protein | | Authors: | Wawrzak, Z, Stogios, P.J, Minasov, G, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | 1.5 Angstrom crystal structure of soluble domain of membrane-anchored thioredoxin family protein from Streptococcus pneumoniae strain Canada MDR_19A
To be Published
|
|
5V26
 
 | |
5MX2
 
 | | Photosystem II depleted of the Mn4CaO5 cluster at 2.55 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Zhang, M, Bommer, M, Chatterjee, R, Hussain, R, Kern, J, Yano, J, Dau, H, Dobbek, H, Zouni, A. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structural insights into the light-driven auto-assembly process of the water-oxidizing Mn4CaO5-cluster in photosystem II.
Elife, 6, 2017
|
|
8IWH
 
 | | Structure and characteristics of a photosystem II supercomplex containing monomeric LHCX and dimeric FCPII antennae from the diatom Thalassiosira pseudonana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Feng, Y, Li, Z.H, Wang, W.D, Shen, J.R. | | Deposit date: | 2023-03-30 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
6DQ5
 
 | |
5ZLG
 
 | | Human duodenal cytochrome b (Dcytb) in zinc ion and ascorbate bound form | | Descriptor: | ASCORBIC ACID, Cytochrome b reductase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ganasen, M, Togashi, H, Mauk, G.A, Shiro, Y, Sawai, H, Sugimoto, H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate.
Commun Biol, 1, 2018
|
|
5ZLE
 
 | | Human duodenal cytochrome b (Dcytb) in substrate free form | | Descriptor: | Cytochrome b reductase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ganasen, M, Togashi, H, Mauk, G.A, Shiro, Y, Sawai, H, Sugimoto, H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate.
Commun Biol, 1, 2018
|
|
5ZP1
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 9 at 288 K (1) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.669 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZW7
 
 | | FAD-PigA complex at 1.3 A | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Lee, C.-C, Ko, T.-P, Wang, A.H.J. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of PigA: A Prolyl Thioester-Oxidizing Enzyme in Prodigiosin Biosynthesis.
Chembiochem, 20, 2019
|
|
2WJ3
 
 | |
6DQB
 
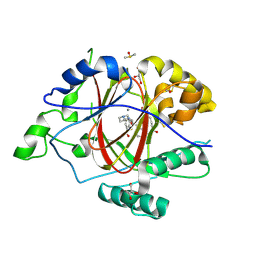 | | LINKED KDM5A JMJ DOMAIN FORMING COVALENT BOND TO INHIBITOR N71 i.e. 2-((3-(4-(dimethylamino)but-2-enamido)phenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 2-{(R)-(3-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(R)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(S)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6BGU
 
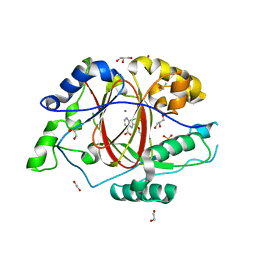 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine (Compound N9) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl)(propoxy)methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
1NSD
 
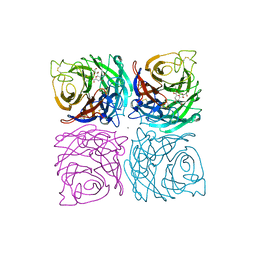 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
1NSC
 
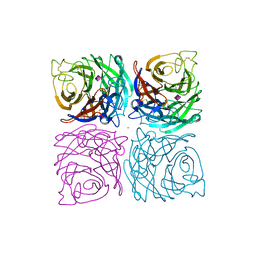 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
6BH0
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (R)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N51) | | Descriptor: | 2-{(R)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
1ODN
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN-EXPOSED PRODUCT FROM ANAEROBIC AC-VINYLGLYCINE FE COMPLEX) | | Descriptor: | 6-(5-AMINO-5-CARBOXY-PENTANOYLAMINO)-3-HYDROXYMETHYL-7-OXO-4-THIA-1-AZA-BICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Elkins, J.M, Rutledge, P.J, Burzlaff, N.I, Clifton, I.J, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Studies on the Reaction of Isopenicillin N Synthase with an Unsaturated Substrate Analogue
Org.Biomol.Chem., 1, 2003
|
|
1LIN
 
 | | CALMODULIN COMPLEXED WITH TRIFLUOPERAZINE (1:4 COMPLEX) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CALMODULIN | | Authors: | Vandonselaar, M, Hickie, R.A, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trifluoperazine-induced conformational change in Ca(2+)-calmodulin.
Nat.Struct.Biol., 1, 1994
|
|
6BGX
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)((4,4-difluorocyclohexyl)methoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N42) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[(4,4-difluorocyclohexyl)methoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
4I3P
 
 | |
