2J31
 
 | | The Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of(Pro)caspase-3 | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CASPASE-3 | | Authors: | Feeney, B, Pop, C, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)Caspase-3.
Biochemistry, 45, 2006
|
|
2WCQ
 
 | | Crystal Structure of Tip-Alpha N34 (HP0596) from Helicobacter pylori at pH4 | | Descriptor: | CITRIC ACID, TNF-ALPHA INDUCER PROTEIN | | Authors: | Tosi, T, Cioci, G, Jouravleva, K, Dian, C, Terradot, L. | | Deposit date: | 2009-03-13 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the Tumor Necrosis Factor Alpha Inducing Protein Tipalpha: A Novel Virulence Factor from Helicobacter Pylori.
FEBS Lett., 583, 2009
|
|
2J32
 
 | | The Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of(Pro)caspase-3 | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CASPASE-3 | | Authors: | Feeney, B, Pop, C, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)Caspase-3.
Biochemistry, 45, 2006
|
|
2ILA
 
 | |
2J30
 
 | | The Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)caspase-3 | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CASPASE-3 | | Authors: | Feeney, B, Pop, C, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)Caspase-3.
Biochemistry, 45, 2006
|
|
2ERW
 
 | |
2J33
 
 | | The Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)caspase-3 | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CASPASE-3 | | Authors: | Feeney, B, Pop, C, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)Caspase-3.
Biochemistry, 45, 2006
|
|
2IWT
 
 | | Thioredoxin h2 (HvTrxh2) in a mixed disulfide complex with the target protein BASI | | Descriptor: | ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CITRATE ANION, THIOREDOXIN H ISOFORM 2 | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2006-07-04 | | Release date: | 2006-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Target Protein Recognition by the Protein Disulfide Reductase Thioredoxin.
Structure, 14, 2006
|
|
1UVF
 
 | |
1X2I
 
 | | Crystal Structure Of Archaeal Xpf/Mus81 Homolog, Hef From Pyrococcus Furiosus, Helix-hairpin-helix Domain | | Descriptor: | Hef helicase/nuclease | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2005-04-24 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Analyses of an Archaeal XPF/Rad1/Mus81 Nuclease: Asymmetric DNA Binding and Cleavage Mechanisms
STRUCTURE, 13, 2005
|
|
1UVG
 
 | |
1YC6
 
 | |
1Y62
 
 | | A 2.4 crystal structure of conkunitzin-S1, a novel Kunitz-fold cone snail neurotoxin. | | Descriptor: | Conkunitzin-S1, SULFATE ION | | Authors: | Dy, C.Y, Buczek, P, Horvath, M.P. | | Deposit date: | 2004-12-03 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of conkunitzin-S1, a neurotoxin and Kunitz-fold disulfide variant from cone snail.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1YQ4
 
 | | Avian respiratory complex ii with 3-nitropropionate and ubiquinone | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 3-NITROPROPANOIC ACID, Coenzyme Q10, ... | | Authors: | Huang, L, Sun, G, Cobessi, D, Wang, A, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
4BJS
 
 | | Crystal structure of the Rif1 C-terminal domain (Rif1-CTD) from Saccharomyces cerevisiae | | Descriptor: | TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Bunker, R.D, Shi, T, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
1BJR
 
 | | COMPLEX FORMED BETWEEN PROTEOLYTICALLY GENERATED LACTOFERRIN FRAGMENT AND PROTEINASE K | | Descriptor: | CALCIUM ION, LACTOFERRIN, PROTEINASE K | | Authors: | Singh, T.P, Sharma, S, Karthikeyan, S, Betzel, C, Bhatia, K.L. | | Deposit date: | 1998-06-27 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of a complex formed between proteolytically-generated lactoferrin fragment and proteinase K.
Proteins, 33, 1998
|
|
1IIE
 
 | |
3OVO
 
 | |
3PV4
 
 | | Structure of Legionella fallonii DegQ (Delta-PDZ2 variant) | | Descriptor: | CADMIUM ION, DegQ | | Authors: | Wrase, R, Scott, H, Hilgenfeld, R, Hansen, G. | | Deposit date: | 2010-12-06 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Legionella HtrA homologue DegQ is a self-compartmentizing protease that forms large 12-meric assemblies.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3Q1D
 
 | | The B-box domain of Trim54 | | Descriptor: | Tripartite motif-containing protein 54, ZINC ION | | Authors: | Walker, J.R, Yermekbayeva, L, Mackenzie, F, Dong, A, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S. | | Deposit date: | 2010-12-17 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The B-Box Domain of Trim54
To be Published
|
|
3Q48
 
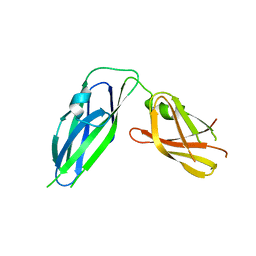 | | Crystal structure of Pseudomonas aeruginosa CupB2 chaperone | | Descriptor: | Chaperone CupB2 | | Authors: | Cai, X, Wang, R, Filloux, A, Waksman, G, Meng, G. | | Deposit date: | 2010-12-23 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of Pseudomonas aeruginosa CupB chaperones
Plos One, 6, 2011
|
|
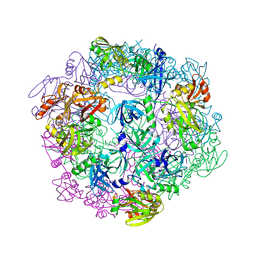
3PV5
 
 | |
3PV3
 
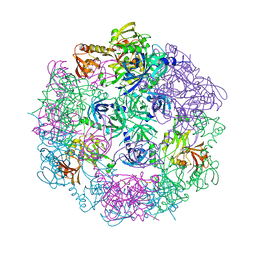 | | Structure of Legionella fallonii DegQ (S193A variant) | | Descriptor: | DegQ, Substrate peptide (Poly-Ala) | | Authors: | Wrase, R, Scott, H, Hilgenfeld, R, Hansen, G. | | Deposit date: | 2010-12-06 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Legionella HtrA homologue DegQ is a self-compartmentizing protease that forms large 12-meric assemblies.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1PVX
 
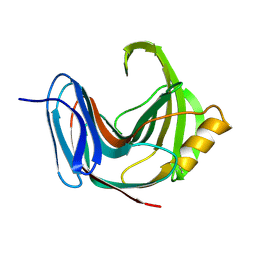 | | DO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.5 | | Descriptor: | PROTEIN (ENDO-1,4-BETA-XYLANASE) | | Authors: | Rajeshkumar, P, Eswaramoorthy, S, Vithayathil, P.J, Viswamitra, M.A. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The tertiary structure at 1.59 A resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti bainier.
J.Mol.Biol., 295, 2000
|
|
3PV2
 
 | |
