3W6S
 
 | | yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BMH
 
 | | Crystal structure of SsHAT | | Descriptor: | ACETYLTRANSFERASE, CHLORIDE ION | | Authors: | He, Y, Turkenburg, J.P, Davies, G.J. | | Deposit date: | 2013-05-08 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-Dimensional Structure of a Streptomyces Sviceus Gnat Acetyltransferase with Similarity to the C-Terminal Domain of the Human Gh84 O-Glcnacase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6SK2
 
 | | HsNMT1 in complex with both MyrCoA and Acetylated-GKSFSKPR peptide reveals N-terminal Lysine Myristoylation | | Descriptor: | Apoptosis-inducing factor 3, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.90000653 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6SK8
 
 | | DeltaC3 C-terminal truncation of HsNMT1 in complex with MyrCoA and GDCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6SJZ
 
 | | HsNMT1 in complex with both MyrCoA and Acetylated-GNCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6ADD
 
 | | The crystal structure of Rv2747 from Mycobacterium tuberculosis in complex with CoA and NLQ | | Descriptor: | Amino-acid acetyltransferase, COENZYME A, N~2~-ACETYL-L-GLUTAMINE | | Authors: | Das, U, Singh, E, Pal, R.K, Tiruttani Subhramanyam, U.K, Gourinath, S, Srinivasan, A. | | Deposit date: | 2018-07-31 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural insights into the substrate binding mechanism of novel ArgA from Mycobacterium tuberculosis
Int. J. Biol. Macromol., 125, 2019
|
|
6SK3
 
 | | C-terminal HsNMT1 deltaC3 truncation in complex with both MyrCoA and GNCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6T9J
 
 | | SAGA Tra1 module | | Descriptor: | Transcription factor SPT20, Transcription initiation factor TFIID subunit 12, Transcription-associated protein 1 | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9I
 
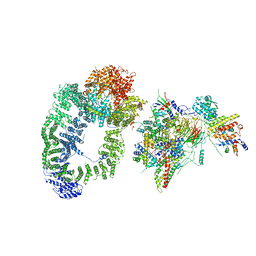 | | cryo-EM structure of transcription coactivator SAGA | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9K
 
 | | SAGA Core module | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6TB4
 
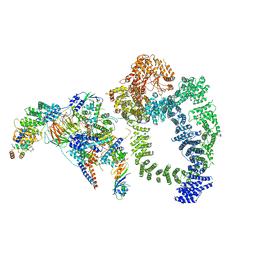 | | Structure of SAGA bound to TBP | | Descriptor: | SAGA-associated factor 73 (Sgf73), Spt20, Subunit (17 kDa) of TFIID and SAGA complexes, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-10-31 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
6BBR
 
 | |
6BC2
 
 | |
6BC7
 
 | | Cryo X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, ACETATE ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC6
 
 | | Cryo X-ray structure of apo AAC-VIa | | Descriptor: | AAC 3-VI protein, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC3
 
 | | Cryo X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, COENZYME A | | Authors: | Cuneo, M.J, Kumar, P, Serpersu, E.H. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC5
 
 | |
6T9L
 
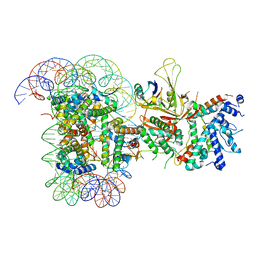 | |
6TBM
 
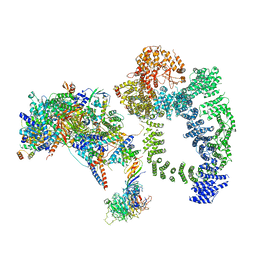 | | Structure of SAGA bound to TBP, including Spt8 and DUB | | Descriptor: | Polyubiquitin-B, SAGA-associated factor 11, Spt20, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-11-01 | | Release date: | 2020-02-12 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
6BBZ
 
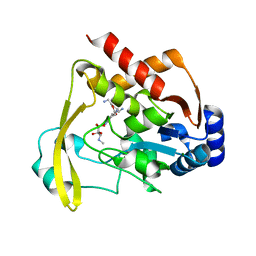 | | Room temperature neutron/X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC4
 
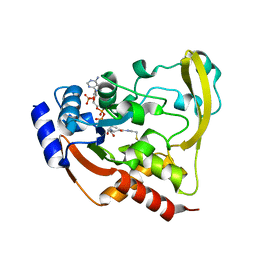 | |
6SKJ
 
 | | DeltaC2 C-terminal truncation of HsNMT1 in complex with MyrCoA and GNCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
5E96
 
 | | Crystal structure of aminoglycoside 6'-acetyltransferase type Ii | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside 6'-acetyltransferase, PHOSPHATE ION | | Authors: | Berghuis, A.M, Burk, D.L, Baettig, O.M, Shi, K. | | Deposit date: | 2015-10-14 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive characterization of ligand-induced plasticity changes in a dimeric enzyme.
Febs J., 283, 2016
|
|
5GIG
 
 | | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Wu, C.Y, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-23 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine
To Be Published
|
|
5GI7
 
 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-phenylethylamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
