2D5F
 
 | | Crystal Structure of Recombinant Soybean Proglycinin A3B4 subunit, its Comparison with Mature Glycinin A3B4 subunit, Responsible for Hexamer Assembly | | Descriptor: | CARBONATE ION, MAGNESIUM ION, glycinin A3B4 subunit | | Authors: | Itoh, T, Adachi, M, Masuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2005-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
2D5G
 
 | |
2D5H
 
 | | Crystal Structure of Recombinant Soybean Proglycinin A3B4 subunit, its Comparison with Mature Glycinin A3B4 subunit, Responsible for Hexamer Assembly | | Descriptor: | CARBONATE ION, MAGNESIUM ION, glycinin A3B4 subunit | | Authors: | Itoh, T, Adachi, M, Masuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2005-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
2D5I
 
 | |
2D5J
 
 | |
2D5K
 
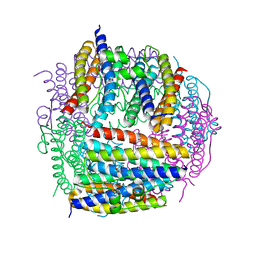 | | Crystal structure of Dps from Staphylococcus aureus | | Descriptor: | Dps family protein, GLYCEROL | | Authors: | Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleoid compaction by MrgA(Asp56Ala/Glu60Ala) does not contribute to staphylococcal cell survival against oxidative stress and phagocytic killing by macrophages
FEMS Microbiol. Lett., 360, 2014
|
|
2D5L
 
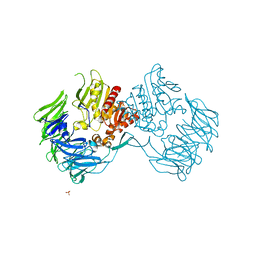 | | Crystal Structure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis | | Descriptor: | SULFATE ION, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2005-11-02 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2D5M
 
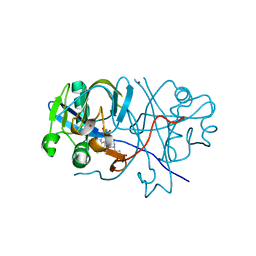 | |
2D5N
 
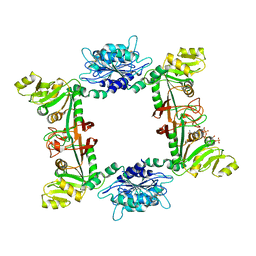 | |
2D5R
 
 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
2D5U
 
 | |
2D5V
 
 | | Crystal structure of HNF-6alpha DNA-binding domain in complex with the TTR promoter | | Descriptor: | 5'-D(*AP*TP*TP*AP*TP*TP*GP*AP*CP*TP*TP*AP*GP*A)-3', 5'-D(*TP*CP*TP*AP*AP*GP*TP*CP*AP*AP*TP*AP*AP*T)-3', ACETATE ION, ... | | Authors: | Iyaguchi, D, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2005-11-07 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6
Structure, 15, 2007
|
|
2D5W
 
 | |
2D5X
 
 | | Crystal structure of carbonmonoxy horse hemoglobin complexed with L35 | | Descriptor: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, CARBON MONOXIDE, Hemoglobin alpha subunit, ... | | Authors: | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | Deposit date: | 2005-11-08 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2D5Y
 
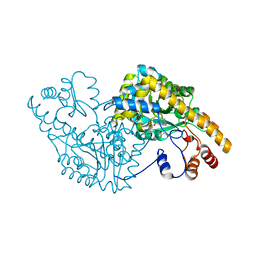 | | Aspartate Aminotransferase Mutant MC With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D5Z
 
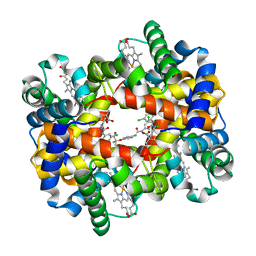 | | Crystal structure of T-state human hemoglobin complexed with three L35 molecules | | Descriptor: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, Hemoglobin alpha subunit, Hemoglobin beta subunit, ... | | Authors: | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | Deposit date: | 2005-11-08 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2D60
 
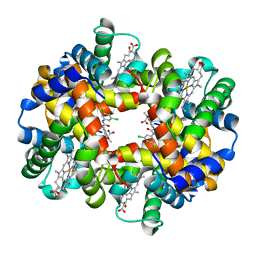 | | Crystal structure of deoxy human hemoglobin complexed with two L35 molecules | | Descriptor: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, Hemoglobin alpha subunit, Hemoglobin beta subunit, ... | | Authors: | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | Deposit date: | 2005-11-08 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2D61
 
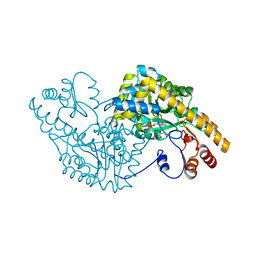 | | Aspartate Aminotransferase Mutant MA With Maleic Acid | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D62
 
 | | Crystal structure of multiple sugar binding transport ATP-binding protein | | Descriptor: | PYROPHOSPHATE 2-, SULFATE ION, multiple sugar-binding transport ATP-binding protein | | Authors: | Lokanath, N.K, Mizohata, E, Yamaguchi-Sihta, E, Chen, L, Liu, Z.J, Wang, B.C, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-08 | | Release date: | 2006-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of multiple sugar binding transport ATP-binding protein
To be Published
|
|
2D63
 
 | | Aspartate Aminotransferase Mutant MA With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D64
 
 | | Aspartate Aminotransferase Mutant MABC With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D65
 
 | | Aspartate Aminotransferase Mutant MABC | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D66
 
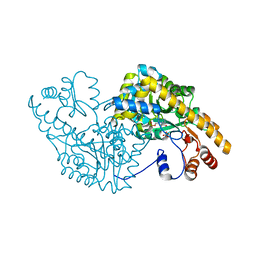 | | Aspartate Aminotransferase Mutant MAB | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D68
 
 | |
2D69
 
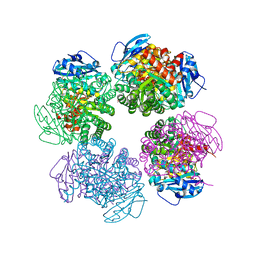 | | Crystal structure of the complex of sulfate ion and octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal) | | Descriptor: | Ribulose bisphosphate carboxylase, SULFATE ION | | Authors: | Mizohata, E, Mishima, C, Akasaka, R, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-10 | | Release date: | 2006-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of sulfate ion and octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal)
To be Published
|
|
