2D34
 
 | | FORMALDEHYDE CROSS-LINKS DAUNORUBICIN AND DNA EFFICIENTLY: HPLC AND X-RAY DIFFRACTION STUDIES | | Descriptor: | 5'-D(*CP*GP*TP*(A35)P*CP*G)-3', DAUNOMYCIN, MAGNESIUM ION | | Authors: | Wang, A.H.-J, Gao, Y.-G, Liaw, Y.-C, Li, Y.-K. | | Deposit date: | 1991-05-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-ray diffraction studies.
Biochemistry, 30, 1991
|
|
2D35
 
 | | Solution structure of Cell Division Reactivation Factor, CedA | | Descriptor: | Cell division activator cedA | | Authors: | Abe, Y, Watanabe, N, Matsuda, Y, Yoshida, Y, Katayama, T, Ueda, T. | | Deposit date: | 2005-09-26 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis and Molecular Interaction of Cell Division Reactivation Factor, CedA from Escherichia coli
To be Published
|
|
2D36
 
 | | The Crystal Structure of Flavin Reductase HpaC | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D37
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NAD+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D38
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NADP+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D39
 
 | | Trivalent Recognition Unit of Innate Immunity System; Crystal Structure of human M-ficolin Fibrinogen-like Domain | | Descriptor: | CALCIUM ION, Ficolin-1 | | Authors: | Tanio, M, Kondo, S, Sugio, S, Kohno, T. | | Deposit date: | 2005-09-26 | | Release date: | 2006-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trivalent recognition unit of innate immunity system; crystal structure of trimeric human m-ficolin fibrinogen-like domain
J.Biol.Chem., 282, 2007
|
|
2D3A
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with ADP and Methionine sulfoximine Phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
2D3B
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with AMPPNP and Methionine sulfoximine | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
2D3C
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with ADP and Phosphinothricin Phosphate | | Descriptor: | (2S)-2-AMINO-4-[METHYL(PHOSPHONOOXY)PHOSPHORYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
2D3D
 
 | | crystal structure of the RNA binding SAM domain of saccharomyces cerevisiae Vts1 | | Descriptor: | CALCIUM ION, Vts1 protein | | Authors: | Aviv, T, Amborski, A.N, Zhao, X.S, Kwan, J.J, Johnson, P.E, Sicheri, F, Donaldson, L.W. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The NMR and X-ray Structures of the Saccharomyces cerevisiae Vts1 SAM Domain Define a Surface for the Recognition of RNA Hairpins
J.Mol.Biol., 356, 2006
|
|
2D3E
 
 | |
2D3F
 
 | | Crystal structures of collagen model peptides (Pro-Pro-Gly)4-Pro-Hyp-Gly-(Pro-Pro-Gly)4 | | Descriptor: | COLLAGEN MODEL PEPTIDES (PRO-PRO-GLY)4-PRO-HYP-GLY-(PRO-PRO-GLY)4 | | Authors: | Wu, G, Noguchi, K, Okuyama, K, Ebisuzaki, S, Nishino, N. | | Deposit date: | 2005-09-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | High-resolution structures of collagen-like peptides [(Pro-Pro-Gly)(4)-Xaa-Yaa-Gly-(Pro-Pro-Gly)(4)]: Implications for triple-helix hydration and Hyp(X) puckering.
Biopolymers, 91, 2009
|
|
2D3G
 
 | | Double sided ubiquitin binding of Hrs-UIM | | Descriptor: | ubiquitin, ubiquitin interacting motif from hepatocyte growth factor-regulated tyrosine kinase substrate | | Authors: | Hirano, S, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2005-09-28 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting
Nat.Struct.Mol.Biol., 13, 2006
|
|
2D3H
 
 | | Crystal structures of collagen model peptides (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 | | Descriptor: | COLLAGEN MODEL PEPTIDES (PRO-PRO-GLY)4-HYP-HYP-GLY-(PRO-PRO-GLY)4 | | Authors: | Wu, G, Noguchi, K, Okuyama, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2005-09-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High-resolution structures of collagen-like peptides [(Pro-Pro-Gly)(4)-Xaa-Yaa-Gly-(Pro-Pro-Gly)(4)]: Implications for triple-helix hydration and Hyp(X) puckering.
Biopolymers, 91, 2009
|
|
2D3I
 
 | | Crystal Structure of Aluminum-Bound Ovotransferrin at 2.15 Angstrom Resolution | | Descriptor: | ALUMINUM ION, BICARBONATE ION, Ovotransferrin | | Authors: | Mizutani, K, Mikami, B, Aibara, S, Hirose, M. | | Deposit date: | 2005-09-28 | | Release date: | 2005-11-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of aluminium-bound ovotransferrin at 2.15 Angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2D3J
 
 | |
2D3K
 
 | |
2D3L
 
 | | Crystal structure of maltohexaose-producing amylase from Bacillus sp.707 complexed with maltopentaose. | | Descriptor: | CALCIUM ION, Glucan 1,4-alpha-maltohexaosidase, SODIUM ION, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2005-09-29 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Trp140 at subsite -6 on the maltohexaose production of maltohexaose-producing amylase from alkalophilic Bacillus sp.707
Protein Sci., 15, 2006
|
|
2D3M
 
 | | Pentaketide chromone synthase complexed with coenzyme A | | Descriptor: | COENZYME A, pentaketide chromone synthase | | Authors: | Morita, H, Kondo, S, Oguro, S, Noguchi, H, Sugio, S, Abe, I, Kohno, T. | | Deposit date: | 2005-09-29 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insight into Chain-Length Control and Product Specificity of Pentaketide Chromone Synthase from Aloe arborescens
Chem.Biol., 14, 2007
|
|
2D3N
 
 | | Crystal structure of maltohexaose-producing amylase from Bacillus sp.707 complexed with maltohexaose | | Descriptor: | CALCIUM ION, Glucan 1,4-alpha-maltohexaosidase, SODIUM ION, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2005-09-29 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Trp140 at subsite -6 on the maltohexaose production of maltohexaose-producing amylase from alkalophilic Bacillus sp.707
Protein Sci., 15, 2006
|
|
2D3O
 
 | | Structure of Ribosome Binding Domain of the Trigger Factor on the 50S ribosomal subunit from D. radiodurans | | Descriptor: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, ... | | Authors: | Schluenzen, F, Wilson, D.N, Hansen, H.A, Tian, P, Harms, J.M, McInnes, S.J, Albrecht, R, Buerger, J, Wilbanks, S.M, Fucini, P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Binding Mode of the Trigger Factor on the Ribosome: Implications for Protein Folding and SRP Interaction
Structure, 13, 2005
|
|
2D3P
 
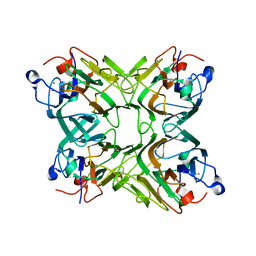 | |
2D3Q
 
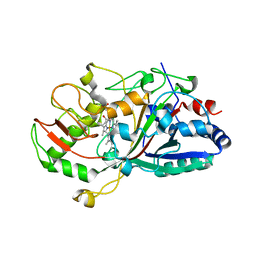 | |
2D3R
 
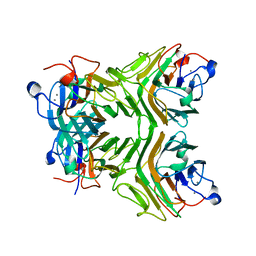 | | Cratylia folibunda seed lectin at acidic pH | | Descriptor: | CALCIUM ION, Lectin alpha chain, MANGANESE (II) ION | | Authors: | Del Sol, F.G, Cavada, B.S, Calvete, J.J. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Cratylia floribunda seed lectin at acidic and basic pHs. Insights into the structural basis of the pH-dependent dimer-tetramer transition.
J.Struct.Biol., 158, 2007
|
|
2D3S
 
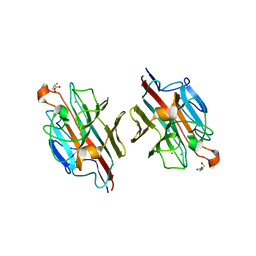 | | Crystal Structure of basic winged bean lectin with Tn-antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, ... | | Authors: | Kulkarni, K.A, Sinha, S, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2005-10-01 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the specificity of basic winged bean lectin for the Tn-antigen: a crystallographic, thermodynamic and modelling study
Febs Lett., 579, 2005
|
|
