7SD6
 
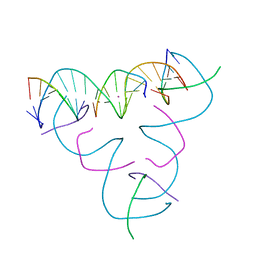 | | [T:Hg2+:T] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SD8
 
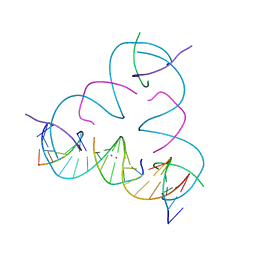 | | [U:Hg2+:U] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*UP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDK
 
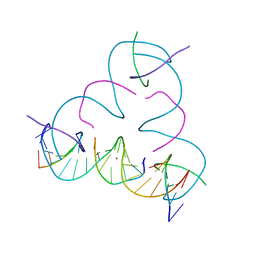 | | [mC:Ag+:S] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5CM)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.86 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDY
 
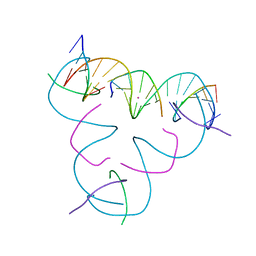 | | [C:Hg2+:U] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDI
 
 | | [T:Hg2+:U] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SE4
 
 | | [iU:Hg2+:C] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5IU)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*CP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.65 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SGB
 
 | | [C-C] DNA mismatch in a self-assembling rhombohedral lattice at pH 5.5 | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*CP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Sha, R, Seeman, N.C. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDH
 
 | | [S:Ag+:S] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(IMC)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDW
 
 | | [T:Hg2+:mC] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(5CM)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SG8
 
 | | [T-T] DNA mismatch in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Bernfeld, W, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SG9
 
 | | [mC-mC] DNA mismatch in a self-assembling rhombohedral lattice at pH 5.5 | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5CM)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(5CM)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SGA
 
 | | [C-S] DNA mismatch in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Bernfeld, W, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
2IDR
 
 | | Crystal structure of translation initiation factor EIF4E from wheat | | Descriptor: | Eukaryotic translation initiation factor 4E-1 | | Authors: | Monzingo, A.F, Sadow, J, Dhaliwal, S, Lyon, A, Hoffman, D.W, Robertus, J.D, Browning, K.S. | | Deposit date: | 2006-09-15 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Plant Physiol., 143, 2007
|
|
4V8Z
 
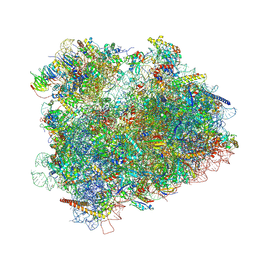 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
6TT9
 
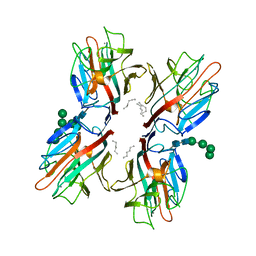 | | rTBL Recombinant Lectin From Tepary Bean | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEXANE, ... | | Authors: | Martinez Alarcon, D, Varrot, A. | | Deposit date: | 2019-12-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recombinant Lectin from Tepary Bean (Phaseolus acutifolius) with Specific Recognition for Cancer-Associated Glycans: Production, Structural Characterization, and Target Identification.
Biomolecules, 10, 2020
|
|
7OZQ
 
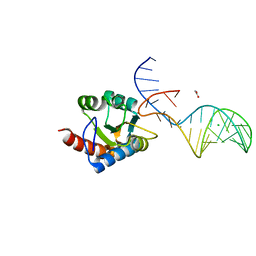 | | Crystal structure of archaeal L7Ae bound to eukaryotic kink-loop | | Descriptor: | 50S ribosomal protein L7Ae, ACETATE ION, CALCIUM ION, ... | | Authors: | Hoefler, S, Lukat, P, Carlomagno, T, Blankenfeldt, W. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Eukaryotic Box C/D methylation machinery has two non-symmetric protein assembly sites.
Sci Rep, 11, 2021
|
|
4GF9
 
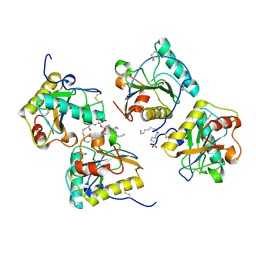 | | Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Yadav, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid.
Plos One, 8, 2013
|
|
6TPY
 
 | |
6TPX
 
 | |
6TQ2
 
 | |
6TPZ
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 5-(1-(1,3-dimethoxypropan-2-yl)-5-morpholino-1H-benzo[d]imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-(1,3-dimethoxypropan-2-yl)-5-morpholin-4-yl-benzimidazol-2-yl]-1,3-dimethyl-pyridin-2-one, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2019-12-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Discovery of a Bromodomain and Extraterminal Inhibitor with a Low Predicted Human Dose through Synergistic Use of Encoded Library Technology and Fragment Screening.
J.Med.Chem., 63, 2020
|
|
6TQ1
 
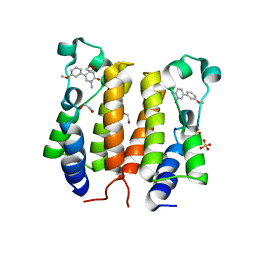 | |
8GM4
 
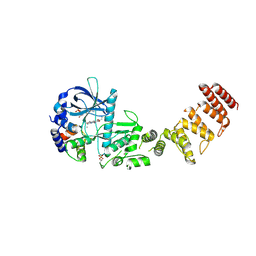 | | Functional construct of the Eukaryotic elongation factor 2 kinase bound to an ATP-competitive inhibitor | | Descriptor: | 7-amino-1-cyclopropyl-3-ethyl-2,4-dioxo-1,2,3,4-tetrahydropyrido[2,3-d]pyrimidine-6-carboxamide, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Piserchio, A, Isiorho, E.A, Dalby, K.N, Ghose, R. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of the complex between calmodulin and a functional construct of eukaryotic elongation factor 2 kinase bound to an ATP-competitive inhibitor.
J.Biol.Chem., 299, 2023
|
|
8GM5
 
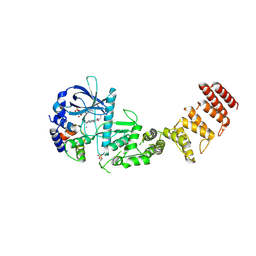 | | Functional construct of the Eukaryotic elongation factor 2 kinase bound to Calmodulin, ADP and to the A-484954 inhibitor and showing two conformations for the 498-520 loop | | Descriptor: | 7-amino-1-cyclopropyl-3-ethyl-2,4-dioxo-1,2,3,4-tetrahydropyrido[2,3-d]pyrimidine-6-carboxamide, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Piserchio, A, Isiorho, E.A, Dalby, K.N, Ghose, R. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of the complex between calmodulin and a functional construct of eukaryotic elongation factor 2 kinase bound to an ATP-competitive inhibitor.
J.Biol.Chem., 299, 2023
|
|
6RE3
 
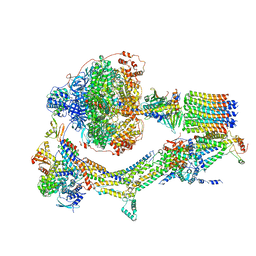 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2B, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
