1ZFA
 
 | | GGA Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*TP*CP*CP*GP*GP*AP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZF8
 
 | | GGT Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*AP*CP*CP*GP*GP*TP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2CD1
 
 | |
5ZDS
 
 | | Crystal structure of the second PDZ domain of Frmpd2 | | Descriptor: | FERM and PDZ domain-containing 2 | | Authors: | Lu, X, Wang, X.F. | | Deposit date: | 2018-02-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The second PDZ domain of scaffold protein Frmpd2 binds to GluN2A of NMDA receptors.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
2CD6
 
 | |
2C83
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 | | Descriptor: | HYPOTHETICAL PROTEIN PM0188 | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
2CD5
 
 | |
2CD3
 
 | |
2C84
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 WITH CMP | | Descriptor: | ALPHA-2,3/2,6-SIALYLTRANSFERASE/SIALIDASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
1ZF0
 
 | | B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*AP*AP*CP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFH
 
 | | TTA Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*TP*AP*AP*TP*TP*AP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2OYC
 
 | | Crystal structure of human pyridoxal phosphate phosphatase | | Descriptor: | Pyridoxal phosphate phosphatase, SODIUM ION, TUNGSTATE(VI)ION | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P69
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with PLP | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
3G7C
 
 | | Structure of the Phosphorylation Mimetic of Occludin C-term Tail | | Descriptor: | Occludin | | Authors: | Tash, B.R, Sundstrom, J.M, Murakami, T, Flanagan, J.M, Bewley, M.C, Antonetii, D.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and analysis of occludin phosphosites: a combined mass spectrometry and bioinformatics approach.
J.PROTEOME RES., 8, 2009
|
|
2KXS
 
 | | ZO1 ZU5 domain in complex with GRINL1A peptide | | Descriptor: | Tight junction protein ZO-1,Myocardial zonula adherens protein | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-30 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Cdc42-dependent formation of the ZO-1/MRCKb complex at the leading edge controls cell migration
Embo J., 30, 2011
|
|
7UMD
 
 | |
2P27
 
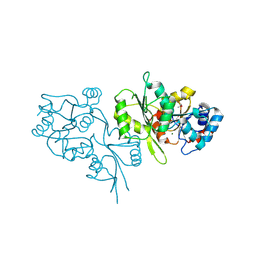 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with Mg2+ at 1.9 A resolution | | Descriptor: | MAGNESIUM ION, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2PVV
 
 | |
2PVW
 
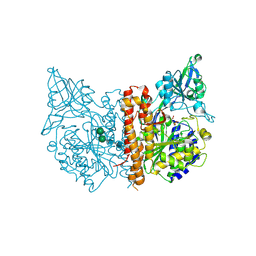 | |
3DFZ
 
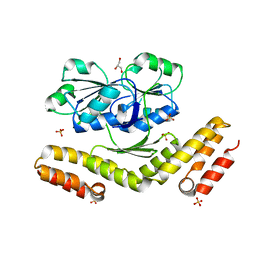 | | SirC, precorrin-2 dehydrogenase | | Descriptor: | GLYCEROL, Precorrin-2 dehydrogenase, SULFATE ION | | Authors: | Schubert, H.L, Hill, C.P, Warren, M.J. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of SirC from Bacillus megaterium: a metal-binding precorrin-2 dehydrogenase
Biochem.J., 415, 2008
|
|
3IFQ
 
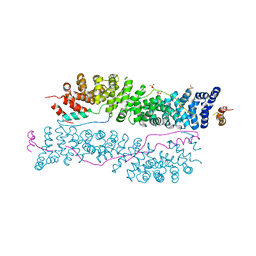 | | Interction of plakoglobin and beta-catenin with desmosomal cadherins | | Descriptor: | E-cadherin, SULFATE ION, plakoglobin | | Authors: | Choi, H.-J, Gross, J.C, Pokutta, S, Weis, W.I. | | Deposit date: | 2009-07-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interactions of plakoglobin and beta-catenin with desmosomal cadherins: basis of selective exclusion of alpha- and beta-catenin from desmosomes.
J.Biol.Chem., 284, 2009
|
|
2USN
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THIADIAZOLE INHIBITOR PNU-141803 | | Descriptor: | CALCIUM ION, STROMELYSIN-1, ZINC ION, ... | | Authors: | Finzel, B.C, Bryant Junior, G.L, Baldwin, E.T. | | Deposit date: | 1998-06-09 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterizations of nonpeptidic thiadiazole inhibitors of matrix metalloproteinases reveal the basis for stromelysin selectivity.
Protein Sci., 7, 1998
|
|
2OR4
 
 | | A high resolution crystal structure of human glutamate carboxypeptidase II in complex with quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Lubkowski, J. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insight into the pharmacophore pocket of human glutamate carboxypeptidase II.
J.Med.Chem., 50, 2007
|
|
7UHY
 
 | | Human GATOR2 complex | | Descriptor: | GATOR complex protein MIOS, GATOR complex protein WDR24, GATOR complex protein WDR59, ... | | Authors: | Rogala, K.B, Valenstein, M.L, Lalgudi, P.V. | | Deposit date: | 2022-03-27 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure of the nutrient-sensing hub GATOR2.
Nature, 607, 2022
|
|
3E5F
 
 | | Crystal Structures of the SMK box (SAM-III) Riboswitch with Se-SAM | | Descriptor: | SMK box (SAM-III) Riboswitch for RNA, STRONTIUM ION, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
