1IHF
 
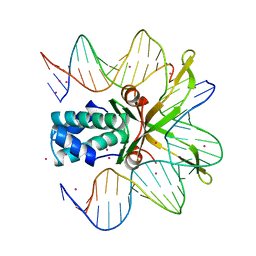 | | INTEGRATION HOST FACTOR/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (35-MER), DNA (5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*C P*AP*CP*C)-3'), ... | | Authors: | Rice, P.A, Yang, S.-W, Mizuuchi, K, Nash, H.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn.
Cell(Cambridge,Mass.), 87, 1996
|
|
8DR6
 
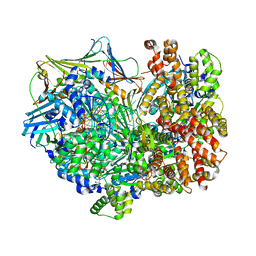 | | Closed state of RFC:PCNA bound to a nicked dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (32-MER), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
5LGY
 
 | |
1FYK
 
 | |
5CBY
 
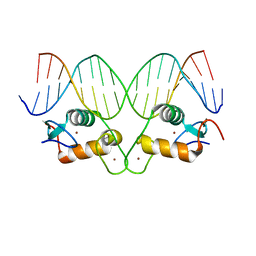 | | AncGR2 DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncGR2 DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CBZ
 
 | | AncMR DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncMR DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8DQW
 
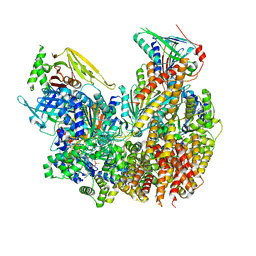 | |
2CGP
 
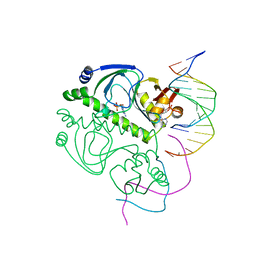 | | CATABOLITE GENE ACTIVATOR PROTEIN/DNA COMPLEX, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*AP*AP*TP*GP*TP*GP*AP*CP*AP*TP*AP*T)-3'), DNA (5'-D(*GP*TP*CP*AP*CP*AP*TP*TP*AP*AP*T)-3'), ... | | Authors: | Passner, J.M, Steitz, T.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
9GSO
 
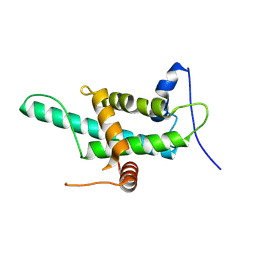 | |
1JJ4
 
 | | Human papillomavirus type 18 E2 DNA-binding domain bound to its DNA target | | Descriptor: | E2 binding site DNA, Regulatory protein E2 | | Authors: | Kim, S.-S, Tam, J.K, Wang, A.F, Hegde, R.S. | | Deposit date: | 2001-07-03 | | Release date: | 2001-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of DNA target discrimination by papillomavirus E2 proteins.
J.Biol.Chem., 275, 2000
|
|
1GA5
 
 | | CRYSTAL STRUCTURE OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB(ALPHA) DNA-BINDING DOMAIN BOUND TO ITS COGNATE RESPONSE ELEMENT | | Descriptor: | 5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*TP*AP*GP*GP*TP*CP*AP*G)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*(5IT)P*G)-3', ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Sierk, M.L, Zhao, Q, Rastinejad, F. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA Deformability as a Recognition Feature in the RevErb Response Element
Biochemistry, 40, 2001
|
|
5J2A
 
 | |
5J2F
 
 | |
5J2D
 
 | |
5J2B
 
 | |
5J2H
 
 | |
7RBG
 
 | | Human DNA polymerase beta crosslinked ternary complex 1 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, CALCIUM ION, ... | | Authors: | Reed, A.J, Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBK
 
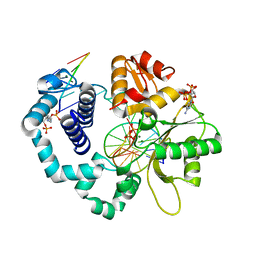 | | Human DNA polymerase beta crosslinked complex, 40 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1EQQ
 
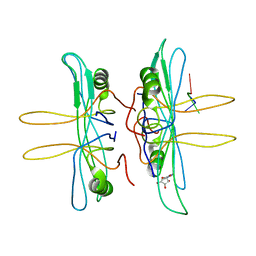 | | SINGLE STRANDED DNA BINDING PROTEIN AND SSDNA COMPLEX | | Descriptor: | 5'-R(*(5MU)P*(5MU)P*(5MU))-3', SINGLE STRANDED DNA BINDING PROTEIN | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Yasuoka, N, Shimamoto, N. | | Deposit date: | 2000-04-06 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis
J.Biochem.(Tokyo), 127, 2000
|
|
1LB2
 
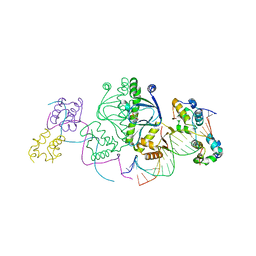 | | Structure of the E. coli alpha C-terminal domain of RNA polymerase in complex with CAP and DNA | | Descriptor: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*AP*G)-3', 5'-D(*CP*TP*TP*TP*TP*TP*TP*CP*CP*TP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Benoff, B, Yang, H, Lawson, C.L, Parkinson, G, Liu, J, Blatter, E, Ebright, Y.W, Berman, H.M, Ebright, R.H. | | Deposit date: | 2002-04-01 | | Release date: | 2002-09-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription activation: the CAP-alpha CTD-DNA complex.
Science, 297, 2002
|
|
4ZTU
 
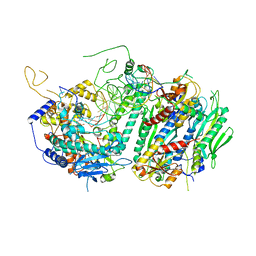 | | Structural basis for processivity and antiviral drug toxicity in human mitochondrial DNA replicase | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (25-MER), DNA (5'-D(P*AP*AP*GP*AP*CP*GP*AP*GP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*GP*TP*AP*C)-3'), ... | | Authors: | Szymanski, M.R, Kuznestov, V.B, Shumate, C.K, Meng, Q, Lee, Y.-S, Patel, G, Patel, S.S, Yin, Y.W. | | Deposit date: | 2015-05-15 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Structural basis for processivity and antiviral drug toxicity
in human mitochondrial DNA replicase
EMBO J., 34, 2015
|
|
5D5U
 
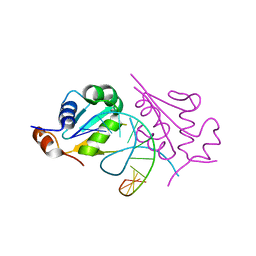 | | Crystal structure of human Hsf1 with HSE DNA | | Descriptor: | Heat shock Element DNA, Heat shock factor protein 1 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7RBE
 
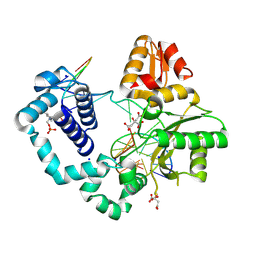 | | Human DNA polymerase beta crosslinked binary complex - A | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, CITRIC ACID, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Reed, A.J, Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBI
 
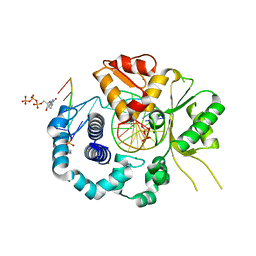 | | Human DNA polymerase beta crosslinked complex, 20 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBO
 
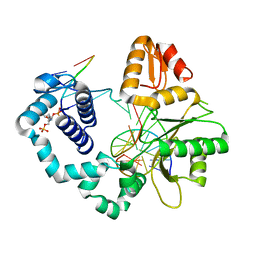 | | Human DNA polymerase beta crosslinked complex, 60 min Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
