1VL5
 
 | |
5W1W
 
 | | Structure of the HLA-E-VMAPRTLVL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
1W7S
 
 | |
1W8X
 
 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
1VLU
 
 | |
1VMG
 
 | |
5W3E
 
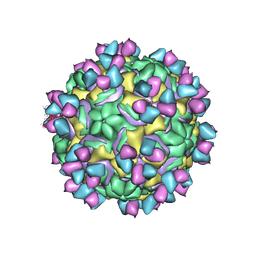 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1VPP
 
 | | COMPLEX BETWEEN VEGF AND A RECEPTOR BLOCKING PEPTIDE | | Descriptor: | PROTEIN (PEPTIDE V108), PROTEIN (VASCULAR ENDOTHELIAL GROWTH FACTOR) | | Authors: | Wiesmann, C, Christinger, H.W, Cochran, A.G, Cunningham, B.C, Fairbrother, W.J, Keenan, C.J, Meng, G, de Vos, A.M. | | Deposit date: | 1998-10-09 | | Release date: | 1999-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex between VEGF and a receptor-blocking peptide.
Biochemistry, 37, 1998
|
|
5W3Y
 
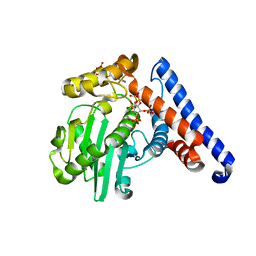 | |
1WAL
 
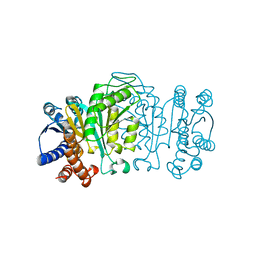 | | 3-ISOPROPYLMALATE DEHYDROGENASE (IPMDH) MUTANT (M219A)FROM THERMUS THERMOPHILUS | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1W3B
 
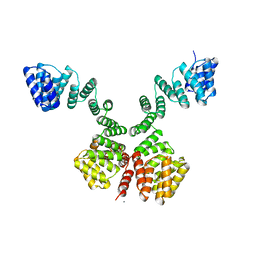 | | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | | Descriptor: | CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYLTRANSFERASE 110 | | Authors: | Jinek, M, Rehwinkel, J, Lazarus, B.D, Izaurralde, E, Hanover, J.A, Conti, E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Superhelical Tpr-Repeat Domain of O-Linked Glcnac Transferase Exhibits Structural Similarities to Importin Alpha
Nat.Struct.Mol.Biol., 11, 2004
|
|
5W4H
 
 | |
7OCC
 
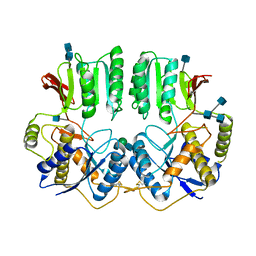 | | NTD of resting state GluA1/A2 heterotertramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 1, ... | | Authors: | Zhang, D, Watson, J.F, Matthews, P.M, Cais, O, Greger, I.H. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Gating and modulation of a hetero-octameric AMPA glutamate receptor.
Nature, 594, 2021
|
|
7OH6
 
 | | Cryo-EM structure of Drs2p-Cdc50p in the [PS]E2-AlFx state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Dieudonne, T, Montigny, C, Boesen, T, Lyons, J.A, Lenoir, G, Nissen, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of substrate-independent phosphorylation in a P4-ATPase lipid flippase
J.Mol.Biol., 2021
|
|
5W4I
 
 | |
1VQ0
 
 | |
1W4G
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state folding transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1VSQ
 
 | |
1VTT
 
 | | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*G)-3') | | Authors: | Ho, P.S, Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG)
Embo J., 4, 1985
|
|
5W5H
 
 | | Human IFIT1 dimer with m7Gppp-AAAA | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1, RNA (5'-D(*(GTA))-R(P*AP*AP*A)-3') | | Authors: | Abbas, Y.M, Martinez-Montero, S, Damha, M.J, Nagar, B. | | Deposit date: | 2017-06-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into IFIT1 dimerization and conformational changes associated with mRNA binding
To Be Published
|
|
5LFU
 
 | |
1VQP
 
 | |
5LG1
 
 | | Room temperature structure of human IgG4-Fc from crystals analysed in situ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-4 chain C region | | Authors: | Davies, A.M, Rispens, T, Ooijevaar-de Heer, P, Aalberse, R.C, Sutton, B.J. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Room temperature structure of human IgG4-Fc from crystals analysed in situ.
Mol. Immunol., 81, 2016
|
|
1VTU
 
 | | Structural characteristics of enantiomorphic DNA: Crystal analysis of racemates of the D(CGCGCG) duplex | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Doi, M, Inoue, M, Tomoo, K, Ishida, T, Ueda, Y, Akagi, M, Urata, H. | | Deposit date: | 1994-08-29 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characteristics of Enantiomorphic DNA: Crystal Analysis of Racemates of the d(CGCGCG) Duplex
J.Am.Chem.Soc., 115, 1993
|
|
1VYQ
 
 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
