2AS4
 
 | | cytochrome c peroxidase in complex with 3-fluorocatechol | | Descriptor: | 3-FLUOROBENZENE-1,2-DIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Brenk, R, Vetter, S.W, Boyce, S.E, Goodin, D.B, Shoichet, B.K. | | Deposit date: | 2005-08-22 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing molecular docking in a charged model binding site.
J.Mol.Biol., 357, 2006
|
|
4BIP
 
 | | Homology model of coxsackievirus A7 (CAV7) full capsid proteins. | | Descriptor: | VP1, VP2, VP3 | | Authors: | Seitsonen, J.J.T, Shakeel, S, Susi, P, Pandurangan, A.P, Sinkovits, R.S, Hyvonen, H, Laurinmaki, P, Yla-Pelto, J, Topf, M, Hyypia, T, Butcher, S.J. | | Deposit date: | 2013-04-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.23 Å) | | Cite: | Combined Approaches to Flexible Fitting and Assessment in Virus Capsids Undergoing Conformational Change.
J.Struct.Biol., 185, 2014
|
|
6HRO
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with inhibitor 118a | | Descriptor: | 1-[2-[4-[4-(4-chlorophenyl)-3-methyl-1~{H}-pyrazol-5-yl]-3-oxidanyl-phenoxy]ethyl]piperidin-1-ium-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ren, J, Zhao, Y, Stuart, D.I. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library.
J.Med.Chem., 62, 2019
|
|
4BDL
 
 | | Crystal structure of the GluK2 K531A LBD dimer in complex with glutamate | | Descriptor: | GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, GLUTAMIC ACID, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4ASE
 
 | | CRYSTAL STRUCTURE OF VEGFR2 (JUXTAMEMBRANE AND KINASE DOMAINS) IN COMPLEX WITH TIVOZANIB (AV-951) | | Descriptor: | TIVOZANIB, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | Authors: | McTigue, M, Deng, Y, Ryan, K, Brooun, A, Diehl, W, Stewart, A. | | Deposit date: | 2012-04-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular Conformations, Interactions, and Properties Associated with Drug Efficiency and Clinical Performance Among Vegfr Tk Inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1R3H
 
 | | Crystal Structure of T10 | | Descriptor: | Beta-2-microglobulin, MHC H2-TL-T10-129 | | Authors: | Rudolph, M.G, Wilson, I.A. | | Deposit date: | 2003-10-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Combined pseudo-merohedral twinning, non-crystallographic symmetry and pseudo-translation in a monoclinic crystal form of the gammadelta T-cell ligand T10.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4BDO
 
 | | Crystal structure of the GluK2 K531A-T779G LBD dimer in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4BDR
 
 | | Crystal structure of the GluK2 R775A LBD dimer in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4BAE
 
 | | Optimisation of pyrroleamides as mycobacterial GyrB ATPase inhibitors: Structure Activity Relationship and in vivo efficacy in the mouse model of tuberculosis | | Descriptor: | 2-[(3S,4R)-4-[(3-bromanyl-4-chloranyl-5-methyl-1H-pyrrol-2-yl)carbonylamino]-3-methoxy-piperidin-1-yl]-4-(2-methyl-1,2,4-triazol-3-yl)-1,3-thiazole-5-carboxylic acid, CALCIUM ION, DNA GYRASE SUBUNIT B, ... | | Authors: | Read, J.A, Gingell, H.G, Madhavapeddi, P. | | Deposit date: | 2012-09-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of Pyrrolamides as Mycobacterial Gyrb ATPase Inhibitors: Structure Activity Relationship and in Vivo Efficacy in the Mouse Model of Tuberculosis.
Antimicrob.Agents Chemother., 58, 2014
|
|
4BDM
 
 | |
4BDQ
 
 | | Crystal structure of the GluK2 R775A LBD dimer in complex with glutamate | | Descriptor: | GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, GLUTAMIC ACID, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4BDN
 
 | | Crystal structure of the GluK2 K531A-T779G LBD dimer in complex with glutamate | | Descriptor: | GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, GLUTAMIC ACID, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
5SV4
 
 | | Anti-Ricin A-chain Single Domain Antibody A3C8 | | Descriptor: | SULFATE ION, Single Domain Antibody A3C8 | | Authors: | Compton, J.R, Legler, P.M. | | Deposit date: | 2016-08-04 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Stability of isolated antibody-antigen complexes as a predictive tool for selecting toxin neutralizing antibodies.
MAbs, 9, 2017
|
|
6I37
 
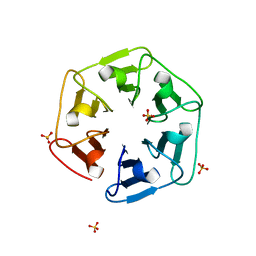 | | Crystal structure of nv1Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | SULFATE ION, nv1Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6I39
 
 | | Crystal structure of v31Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | MAGNESIUM ION, v31Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
1RFD
 
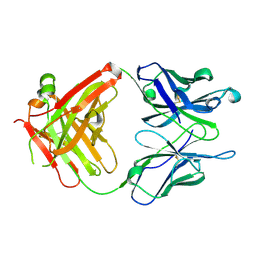 | | ANTI-COCAINE ANTIBODY M82G2 | | Descriptor: | Fab M82G2, Heavy Chain, Light Chain | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-11-07 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Diversity in hapten recognition: structural study of an anti-cocaine antibody M82G2.
J.Mol.Biol., 349, 2005
|
|
6I38
 
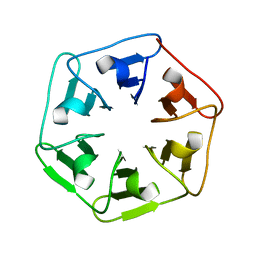 | |
1RIH
 
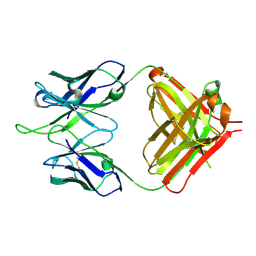 | | Crystal Structure of Fab 14F7, a unique anti-tumor antibody specific for N-glycolyl GM3 | | Descriptor: | heavy chain of antibody 14F7, light chain of antibody 14F7 | | Authors: | Krengel, U, Olsson, L.-L, Martinez, C, Talavera, A, Rojas, G, Mier, E, Angstrom, J, Moreno, E. | | Deposit date: | 2003-11-17 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Molecular Interactions of a Unique Antitumor Antibody Specific for N-Glycolyl GM3.
J.Biol.Chem., 279, 2004
|
|
5TOG
 
 | | Room temperature structure of ubiquitin variant u7ub25.2540 | | Descriptor: | Polyubiquitin-B, SULFATE ION | | Authors: | Biel, J.T, Thompson, M.C, Cunningham, C.N, Corn, J.E, Fraser, J.S. | | Deposit date: | 2016-10-17 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Flexibility and Design: Conformational Heterogeneity along the Evolutionary Trajectory of a Redesigned Ubiquitin.
Structure, 25, 2017
|
|
1RP1
 
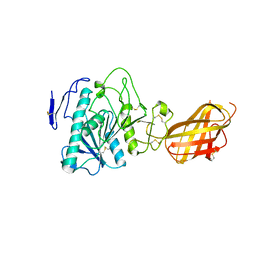 | | DOG PANCREATIC LIPASE RELATED PROTEIN 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PANCREATIC LIPASE RELATED PROTEIN 1 | | Authors: | Roussel, A, Cambillau, C. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reactivation of the totally inactive pancreatic lipase RP1 by structure-predicted point mutations.
Proteins, 32, 1998
|
|
1R8G
 
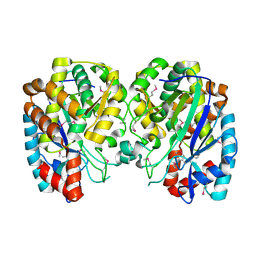 | | Structure and function of YbdK | | Descriptor: | Hypothetical protein ybdK | | Authors: | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-10-24 | | Release date: | 2004-08-17 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
5U3E
 
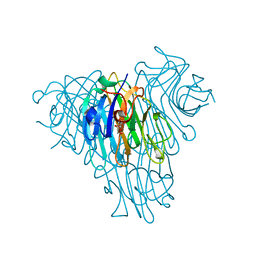 | | Crystal Structure of Native Lectin from Canavalia bonariensis Seeds (CaBo) complexed with alpha-methyl-D-mannoside | | Descriptor: | CALCIUM ION, Canavalia bonariensis seed lectin, MANGANESE (II) ION, ... | | Authors: | Silva, M.T.L, Osterne, V.J.S, Pinto-Junior, V.R, Santiago, M.Q, Araripe, D.A, Neco, A.H.B, Silva-Filho, J.C, Martins, J.L, Rocha, C.R.C, Leal, R.B, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Canavalia bonariensis lectin: Molecular bases of glycoconjugates interaction and antiglioma potential.
Int. J. Biol. Macromol., 106, 2018
|
|
8CYK
 
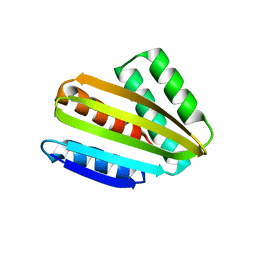 | | Crystal structure of hallucinated protein HALC1_878 | | Descriptor: | HALC1_878 | | Authors: | Ragotte, R.J, Bera, A.K, Milles, L.F, Wicky, B.I.M, Baker, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Robust deep learning-based protein sequence design using ProteinMPNN.
Science, 378, 2022
|
|
5SYA
 
 | |
5SY4
 
 | |
