8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8AWO
 
 | |
8AWP
 
 | |
7LC7
 
 | |
8QMM
 
 | | M291I variant of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Omeiri, J, Martin, L, Usclat, A, Cherrier, M.V, Nicolet, Y. | | Deposit date: | 2023-09-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Maturation of the [FeFe]-Hydrogenase: Direct Transfer of the ( kappa 3 -cysteinate)Fe II (CN)(CO) 2 Complex B from HydG to HydE.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7LC5
 
 | |
6OZF
 
 | |
6OZG
 
 | |
5LQS
 
 | | Structure of quinolinate synthase Y21F mutant in complex with substrate-derived quinolinate | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, QUINOLINIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2016-08-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
3DIN
 
 | | Crystal structure of the protein-translocation complex formed by the SecY channel and the SecA ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Zimmer, J, Nam, Y, Rapoport, T.A. | | Deposit date: | 2008-06-20 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a complex of the ATPase SecA and the protein-translocation channel.
Nature, 455, 2008
|
|
5LAD
 
 | |
4W9M
 
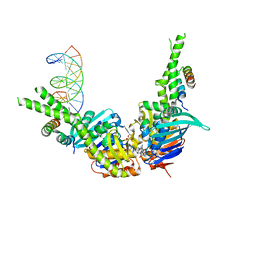 | | AMPPNP bound Rad50 in complex with dsDNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*GP*TP*CP*AP*CP*CP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*GP*GP*TP*GP*AP*CP*CP*GP*AP*CP*C)-3'), Exonuclease, ... | | Authors: | Rojowska, A, Lammens, K. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Rad50 DNA double-strand break repair protein in complex with DNA.
Embo J., 33, 2014
|
|
5B7Y
 
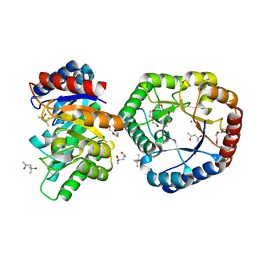 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ketose-3-Epimerase with Co2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, COBALT (II) ION, ... | | Authors: | Cao, T.P, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
5B80
 
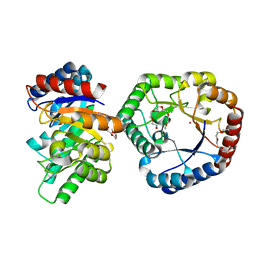 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ketose-3-Epimerase with Cu2+ | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, COPPER (II) ION, Uncharacterized protein TM_0416 | | Authors: | Cao, T.P, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
6Y86
 
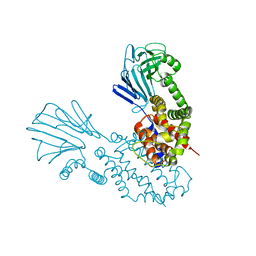 | |
4M8A
 
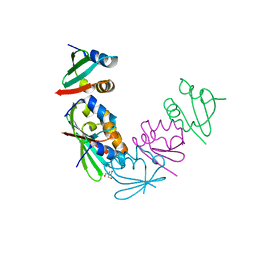 | | Crystal Structure of Thermotoga maritima FtsH Periplasmic Domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-dependent zinc metalloprotease FtsH | | Authors: | An, J.Y, Sharif, H, Barrera, F.N, Karabadzhak, A, Kang, G.B, Park, K.J, Sakkiah, S, Lee, K.W, Lee, S, Engelman, D.M, Wang, J, Eom, S.H. | | Deposit date: | 2013-08-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural roles of periplasmic and transmembrane domains of FtsH in ATP-dependent proteolysis
To be Published
|
|
1B3Q
 
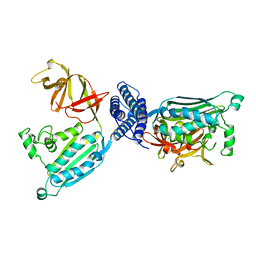 | | CRYSTAL STRUCTURE OF CHEA-289, A SIGNAL TRANSDUCING HISTIDINE KINASE | | Descriptor: | MERCURY (II) ION, PROTEIN (CHEMOTAXIS PROTEIN CHEA) | | Authors: | Bilwes, A.M, Alex, L.A, Crane, B.R, Simon, M.I. | | Deposit date: | 1998-12-14 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of CheA, a signal-transducing histidine kinase.
Cell(Cambridge,Mass.), 96, 1999
|
|
5B7Z
 
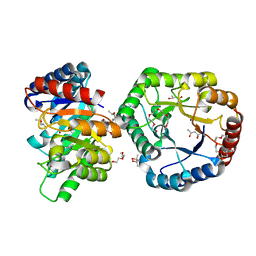 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ketose-3-Epimerase with Ni2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, NICKEL (II) ION, ... | | Authors: | Cao, T.P, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
1I5D
 
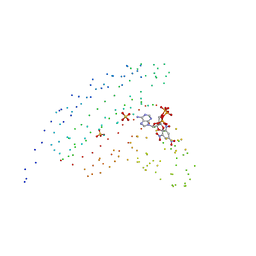 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH TNP-ATP | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE, SULFATE ION | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
4Q0F
 
 | | Crystal Structure of Thermotoga maritima FtsH Periplasmic domain | | Descriptor: | ATP-dependent zinc metalloprotease FtsH | | Authors: | An, J.Y, Sharif, H, Barrera, F.N, Karabadzhak, A, Kang, G.B, Park, K.J, Sakkiah, S, Lee, K.W, Lee, S, Engelman, D.M, Wang, J, Eom, S.H. | | Deposit date: | 2014-04-01 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structures of periplasmic and transmembrane domains of FtsH suggest a reverse translocon mechanism for protein extraction from membrane
To be Published
|
|
4Q37
 
 | |
4P3X
 
 | | Structure of the Fe4S4 quinolinate synthase NadA from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Cherrier, M.V, Chan, A, Darnault, C, Reichmann, D, Amara, P, Ollagnier de Choudens, S, Fontecilla-Camps, J.C. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of Fe4S4 quinolinate synthase unravels an enzymatic dehydration mechanism that uses tyrosine and a hydrolase-type triad.
J.Am.Chem.Soc., 136, 2014
|
|
5F35
 
 | | Structure of quinolinate synthase in complex with citrate | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
5F33
 
 | | Structure of quinolinate synthase in complex with phosphoglycolohydroxamate | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHOGLYCOLOHYDROXAMIC ACID, Quinolinate synthase A, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
7QAZ
 
 | |
