7BR3
 
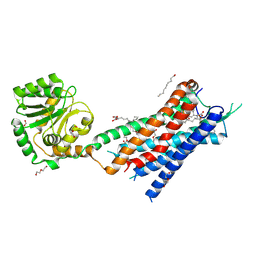 | | Crystal structure of the protein 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | Authors: | Cheng, L, Shao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
2P53
 
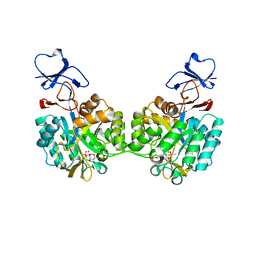 | | Crystal structure of N-acetyl-D-glucosamine-6-phosphate deacetylase D273N mutant complexed with N-acetyl phosphonamidate-d-glucosamine-6-phosphate | | Descriptor: | 2-deoxy-2-{[(S)-hydroxy(methyl)phosphoryl]amino}-6-O-phosphono-alpha-D-glucopyranose, N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Hall, R.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2007-03-14 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Diversity within the Mononuclear and Binuclear Active Sites of N-Acetyl-d-glucosamine-6-phosphate Deacetylase.
Biochemistry, 46, 2007
|
|
8BPP
 
 | |
4Z2R
 
 | |
8AWR
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with L-carbaxylosyl chloride | | Descriptor: | (1~{S},2~{R},3~{S},6~{S})-6-chloranylcyclohex-4-ene-1,2,3-triol, (1~{S},2~{S},3~{S},4~{R})-cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
8AX3
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with L-carbaxylosyl fluoride | | Descriptor: | (1~{S},2~{R},3~{S},6~{S})-6-fluoranylcyclohex-4-ene-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
6D9M
 
 | | T4-Lysozyme fusion to Geobacter GGDEF | | Descriptor: | ACETATE ION, Fusion protein of Endolysin,Response receiver sensor diguanylate cyclase, GAF domain-containing, ... | | Authors: | Hallberg, Z, Doxzen, K, Kranzusch, P.J, Hammond, M. | | Deposit date: | 2018-04-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and mechanism of a Hypr GGDEF enzyme that activates cGAMP signaling to control extracellular metal respiration.
Elife, 8, 2019
|
|
4Z0H
 
 | | X-ray structure of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase (GapC1) complexed with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Orru, R, Falini, G, Trost, P. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tuning Cysteine Reactivity and Sulfenic Acid Stability by Protein Microenvironment in Glyceraldehyde-3-Phosphate Dehydrogenases of Arabidopsis thaliana.
Antioxid. Redox Signal., 24, 2016
|
|
7UZM
 
 | | Glutamate dehydrogenase 1 from human liver | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Zhang, Z. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
6HDH
 
 | |
6QWZ
 
 | | HEWL lysozyme, crystallized from KCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
2P7M
 
 | | Crystal structure of monoclinic form of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes at pH 6.5 | | Descriptor: | Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
6R1N
 
 | | Crystal structure of S. aureus seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
4YZ4
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with N-Acetylneuraminic acid. | | Descriptor: | N-acetyl-alpha-neuraminic acid, SULFATE ION, Sialidase, ... | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
6QY1
 
 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 1500 patterns merged | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
7BYU
 
 | | Crystal structure of Acidovorax avenae L-fucose mutarotase (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, L-fucose mutarotase | | Authors: | Watanabe, Y, Fukui, Y, Watanabe, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Functional and structural characterization of a novel L-fucose mutarotase involved in non-phosphorylative pathway of L-fucose metabolism.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
7BYV
 
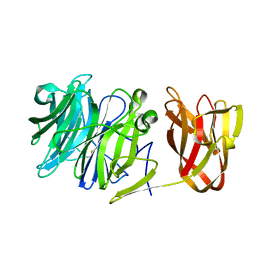 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A E208Q with beta-1,3-galactotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Galactan 1,3-beta-galactosidase, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
7BZK
 
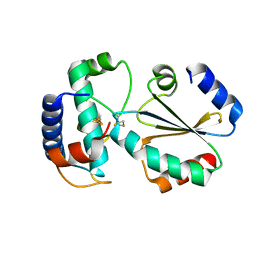 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin y1 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, IRON/SULFUR CLUSTER, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5935 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
2YNG
 
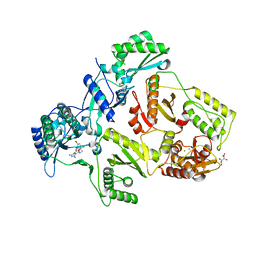 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, MAGNESIUM ION, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
8BPQ
 
 | |
7C2D
 
 | | Esterase AlinE4 mutant-S13A | | Descriptor: | ACETATE ION, CADMIUM ION, SGNH-hydrolase family esterase | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | C-terminal swapped dimers revealed a new catalytic mechanism of SGNH-hydrolase family esterases
To Be Published
|
|
8BH3
 
 | | DNA-PK Ku80 mediated dimer bound to PAXX | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Hardwick, S.W, Chaplin, A.K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
2YNF
 
 | | HIV-1 Reverse Transcriptase Y188L mutant in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
4Z25
 
 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.339 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
8BHV
 
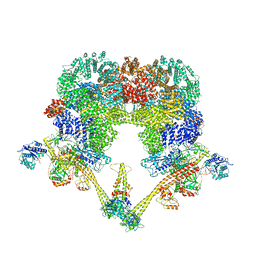 | | DNA-PK XLF mediated dimer bound to PAXX | | Descriptor: | DNA (24-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Hardwick, S.W, Chaplin, A.K. | | Deposit date: | 2022-11-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
