2GLT
 
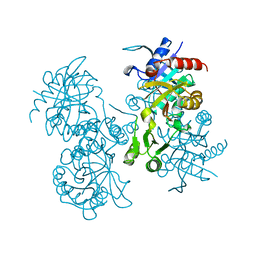 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
1MBE
 
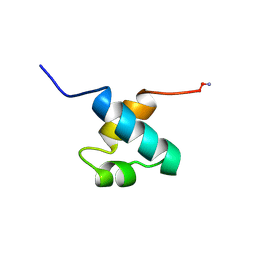 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBK
 
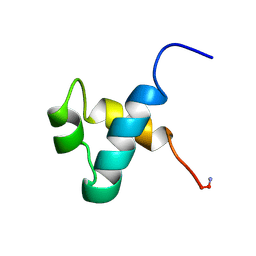 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBG
 
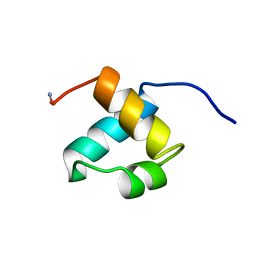 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1HPM
 
 | |
1CSN
 
 | | BINARY COMPLEX OF CASEIN KINASE-1 WITH MGATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CASEIN KINASE-1, MAGNESIUM ION, ... | | Authors: | Xu, R.-M, Cheng, X. | | Deposit date: | 1995-04-25 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of casein kinase-1, a phosphate-directed protein kinase.
EMBO J., 14, 1995
|
|
1CTE
 
 | |
1BNR
 
 | | BARNASE | | Descriptor: | BARNASE (G SPECIFIC ENDONUCLEASE) | | Authors: | Bycroft, M. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional solution structure of barnase using nuclear magnetic resonance spectroscopy.
Biochemistry, 30, 1991
|
|
1CGO
 
 | | CYTOCHROME C' | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Dobbs, A.J, Faber, H.R, Anderson, B.F, Baker, E.N. | | Deposit date: | 1995-05-01 | | Release date: | 1995-07-31 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of cytochrome c' from two Alcaligenes species and the implications for four-helix bundle structures.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1MBF
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
201D
 
 | |
1FIV
 
 | | STRUCTURE OF AN INHIBITOR COMPLEX OF PROTEINASE FROM FELINE IMMUNODEFICIENCY VIRUS | | Descriptor: | FIV PROTEASE, FIV PROTEASE INHIBITOR ACE-ALN-VAL-STA-GLU-ALN-NH2 | | Authors: | Wlodawer, A, Gustchina, A, Reshetnikova, L, Lubkowski, J, Zdanov, A. | | Deposit date: | 1995-05-04 | | Release date: | 1995-07-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an inhibitor complex of the proteinase from feline immunodeficiency virus.
Nat.Struct.Biol., 2, 1995
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1PDN
 
 | | CRYSTAL STRUCTURE OF A PAIRED DOMAIN-DNA COMPLEX AT 2.5 ANGSTROMS RESOLUTION REVEALS STRUCTURAL BASIS FOR PAX DEVELOPMENTAL MUTATIONS | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*TP*CP*AP*CP*GP*GP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*AP*AP*CP*CP*GP*TP*GP*AP*CP*G)-3'), PROTEIN (PRD PAIRED) | | Authors: | Xu, W, Rould, M.A, Jun, S, Desplan, C, Pabo, C.O. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations.
Cell(Cambridge,Mass.), 80, 1995
|
|
1HYL
 
 | | THE 1.8 A STRUCTURE OF COLLAGENASE FROM HYPODERMA LINEATUM | | Descriptor: | HYPODERMA LINEATUM COLLAGENASE | | Authors: | Broutin, I, Arnoux, B, Riche, C, Lecroisey, A, Keil, B, Pascard, C, Ducruix, A. | | Deposit date: | 1995-05-02 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A structure of Hypoderma lineatum collagenase: a member of the serine proteinase family.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1TLF
 
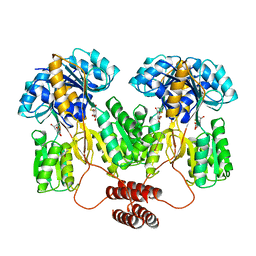 | |
1OPG
 
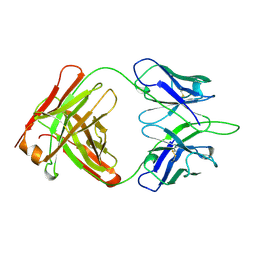 | | OPG2 FAB FRAGMENT | | Descriptor: | OPG2 FAB (HEAVY CHAIN), OPG2 FAB (LIGHT CHAIN) | | Authors: | Kodandapani, R, Veerapandian, B, Ely, K.R. | | Deposit date: | 1995-04-28 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the OPG2 Fab. An antireceptor antibody that mimics an RGD cell adhesion site.
J.Biol.Chem., 270, 1995
|
|
1PVD
 
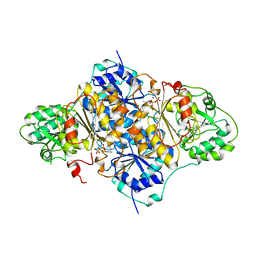 | |
1BVD
 
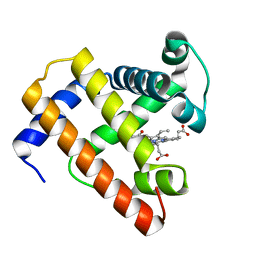 | | STRUCTURE OF A BILIVERDIN APOMYOGLOBIN COMPLEX (FORM B) AT 98 K | | Descriptor: | APOMYOGLOBIN, BILIVERDINE IX ALPHA | | Authors: | Wagner, U.G, Mueller, N, Schmitzberger, W, Falk, H, Kratky, C. | | Deposit date: | 1994-12-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure determination of the biliverdin apomyoglobin complex: crystal structure analysis of two crystal forms at 1.4 and 1.5 A resolution.
J.Mol.Biol., 247, 1995
|
|
1BVC
 
 | | STRUCTURE OF A BILIVERDIN APOMYOGLOBIN COMPLEX (FORM D) AT 118 K | | Descriptor: | APOMYOGLOBIN, BILIVERDINE IX ALPHA, PHOSPHATE ION | | Authors: | Wagner, U.G, Mueller, N, Schmitzberger, W, Falk, H, Kratky, C. | | Deposit date: | 1994-12-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure determination of the biliverdin apomyoglobin complex: crystal structure analysis of two crystal forms at 1.4 and 1.5 A resolution.
J.Mol.Biol., 247, 1995
|
|
1LCP
 
 | |
1PSD
 
 | |
1PNE
 
 | | CRYSTALLIZATION AND STRUCTURE DETERMINATION OF BOVINE PROFILIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | PROFILIN | | Authors: | Cedergren-Zeppezauer, E.S, Goonesekere, N.C.W, Rozycki, M.D, Myslik, J.C, Dauter, Z, Lindberg, U, Schutt, C.E. | | Deposit date: | 1995-05-05 | | Release date: | 1995-07-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and structure determination of bovine profilin at 2.0 A resolution.
J.Mol.Biol., 240, 1994
|
|
1PLP
 
 | | SOLUTION STRUCTURE OF THE CYTOPLASMIC DOMAIN OF PHOSPHOLAMBAN | | Descriptor: | PHOSPHOLAMBAN | | Authors: | Mortishire-Smith, R.J, Pitzenberger, S.M, Burke, C.J, Middaugh, C.R, Garsky, V.M, Johnson, R.G. | | Deposit date: | 1995-05-01 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cytoplasmic domain of phopholamban: phosphorylation leads to a local perturbation in secondary structure.
Biochemistry, 34, 1995
|
|
1POG
 
 | | SOLUTION STRUCTURE OF THE OCT-1 POU-HOMEO DOMAIN DETERMINED BY NMR AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | OCT-1 POU HOMEODOMAIN DNA-BINDING PROTEIN | | Authors: | Cox, M, Van Tilborg, P.J.A, De Laat, W, Boelens, R, Van Leeuwen, H.C, Van Der Vliet, P.C, Kaptein, R. | | Deposit date: | 1994-10-12 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Oct-1 POU homeodomain determined by NMR and restrained molecular dynamics.
J.Biomol.NMR, 6, 1995
|
|
