1CEO
 
 | |
1CEQ
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
1CER
 
 | |
1CES
 
 | | CRYSTALS OF DEMETALLIZED CONCANAVALIN A SOAKED WITH ZINC HAVE A ZINC ION BOUND IN THE S1 SITE | | Descriptor: | CONCANAVALIN A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Poortmans, F, Wyns, L. | | Deposit date: | 1996-02-15 | | Release date: | 1997-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sequential structural changes upon zinc and calcium binding to metal-free concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1CET
 
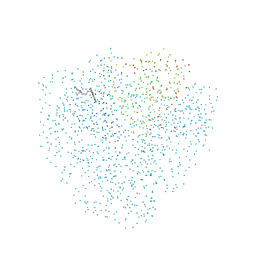 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
1CEU
 
 | |
1CEV
 
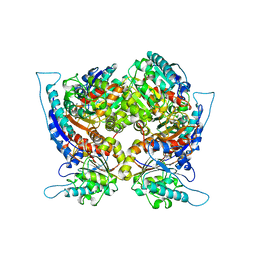 | | ARGINASE FROM BACILLUS CALDOVELOX, NATIVE STRUCTURE AT PH 5.6 | | Descriptor: | MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
1CEW
 
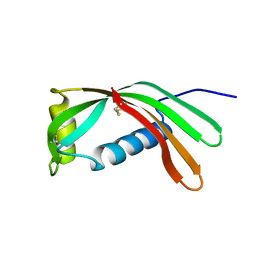 | |
1CEX
 
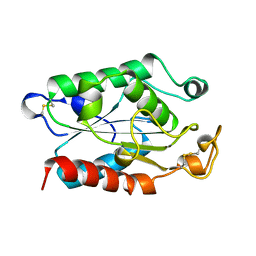 | | STRUCTURE OF CUTINASE | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Czjzek, M, Lamzin, V, Nicolas, A, Cambillau, C. | | Deposit date: | 1997-02-18 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution (1.0 A) crystal structure of Fusarium solani cutinase: stereochemical analysis.
J.Mol.Biol., 268, 1997
|
|
1CEY
 
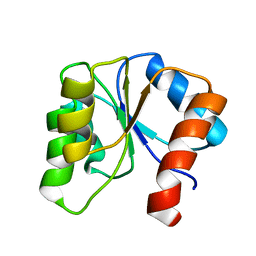 | | ASSIGNMENTS, SECONDARY STRUCTURE, GLOBAL FOLD, AND DYNAMICS OF CHEMOTAXIS Y PROTEIN USING THREE-AND FOUR-DIMENSIONAL HETERONUCLEAR (13C,15N) NMR SPECTROSCOPY | | Descriptor: | CHEY | | Authors: | Moy, F.J, Lowry, D.F, Matsumura, P, Dahlquist, F.W, Krywko, J.E, Domaille, P.J. | | Deposit date: | 1994-11-23 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C,15N) NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CEZ
 
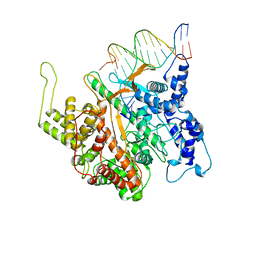 | | CRYSTAL STRUCTURE OF A T7 RNA POLYMERASE-T7 PROMOTER COMPLEX | | Descriptor: | DNA (5'-D(P*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP*AP*TP*TP*A)-3'), PROTEIN (BACTERIOPHAGE T7 RNA POLYMERASE) | | Authors: | Cheetham, G.M.T, Jeruzalmi, D, Steitz, T.A. | | Deposit date: | 1999-03-11 | | Release date: | 1999-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for initiation of transcription from an RNA polymerase-promoter complex.
Nature, 399, 1999
|
|
1CF0
 
 | |
1CF1
 
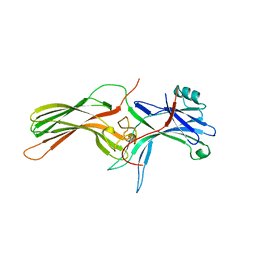 | | ARRESTIN FROM BOVINE ROD OUTER SEGMENTS | | Descriptor: | PROTEIN (ARRESTIN) | | Authors: | Hirsch, J.A, Schubert, C, Gurevich, V.V, Sigler, P.B. | | Deposit date: | 1999-03-23 | | Release date: | 1999-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8 A crystal structure of visual arrestin: a model for arrestin's regulation.
Cell(Cambridge,Mass.), 97, 1999
|
|
1CF2
 
 | | THREE-DIMENSIONAL STRUCTURE OF D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM THE HYPERTHERMOPHILIC ARCHAEON METHANOTHERMUS FERVIDUS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Charron, C, Talfournier, F, Isuppov, M.N, Branlant, G, Littlechild, J.A, Vitoux, B, Aubry, A. | | Deposit date: | 1999-03-24 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallization and preliminary X-ray diffraction studies of D-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Methanothermus fervidus.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CF3
 
 | | GLUCOSE OXIDASE FROM APERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (GLUCOSE OXIDASE), ... | | Authors: | Hecht, H.J, Kalisz, H. | | Deposit date: | 1999-03-23 | | Release date: | 1999-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CF4
 
 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
1CF5
 
 | | BETA-MOMORCHARIN STRUCTURE AT 2.55 A | | Descriptor: | PROTEIN (BETA-MOMORCHARIN), beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yuan, Y.-R, He, Y.-N, Xiong, J.-P, Xia, Z.-X. | | Deposit date: | 1999-03-24 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structure of beta-momorcharin at 2.55 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CF7
 
 | | STRUCTURAL BASIS OF DNA RECOGNITION BY THE HETERODIMERIC CELL CYCLE TRANSCRIPTION FACTOR E2F-DP | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*TP*CP*GP*CP*GP*CP*GP*GP*TP*TP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*CP*CP*GP*CP*GP*CP*GP*AP*AP*AP*A)-3'), PROTEIN (TRANSCRIPTION FACTOR DP-2), ... | | Authors: | Zheng, N, Fraenkel, E, Pabo, C.O, Pavletich, N.P. | | Deposit date: | 1999-03-24 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP.
Genes Dev., 13, 1999
|
|
1CF8
 
 | | Convergence of catalytic antibody and terpene cyclase mechanisms: polyene cyclization directed by carbocation-pi interactions | | Descriptor: | 4-{4-[2-(1A,7A-DIMETHYL-4-OXY-OCTAHYDRO-1-OXA-4-AZA-CYCLOPROPA[A]NAPHTHALEN-4-YL) -ACETYLAMINO]-PHENYLCARBAMOYL}-BUTYRIC ACID, CADMIUM ION, PROTEIN (CATALYTIC ANTIBODY 19A4 (HEAVY CHAIN)), ... | | Authors: | Paschall, C.M, Hasserodt, J, Jones, T, Lerner, R.A, Janda, K.D, Christianson, D.W. | | Deposit date: | 1999-03-24 | | Release date: | 1999-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Convergence of Catalytic Antibody and Terpene Cyclase Mechanisms: Polyene Cyclization Directed by Carbocation-pi Interactions
Angew.Chem.Int.Ed.Engl., 38, 1999
|
|
1CF9
 
 | |
1CFA
 
 | | SOLUTION STRUCTURE OF A SEMI-SYNTHETIC C5A RECEPTOR ANTAGONIST AT PH 5.2, 303K, NMR, 20 STRUCTURES | | Descriptor: | COMPLEMENT 5A SEMI-SYNTHETIC ANTAGONIST, SYNTHETIC N-TERMINAL TAIL | | Authors: | Zhang, X, Boyar, W, Galakatos, N, Gonnella, N.C. | | Deposit date: | 1996-09-21 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a unique C5a semi-synthetic antagonist: implications in receptor binding.
Protein Sci., 6, 1997
|
|
1CFB
 
 | | CRYSTAL STRUCTURE OF TANDEM TYPE III FIBRONECTIN DOMAINS FROM DROSOPHILA NEUROGLIAN AT 2.0 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DROSOPHILA NEUROGLIAN, ... | | Authors: | Huber, A.H, Wang, Y.E, Bieber, A.J, Bjorkman, P.J. | | Deposit date: | 1994-08-27 | | Release date: | 1994-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tandem type III fibronectin domains from Drosophila neuroglian at 2.0 A.
Neuron, 12, 1994
|
|
1CFC
 
 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-08-02 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CFD
 
 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-10-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CFE
 
 | | P14A, NMR, 20 STRUCTURES | | Descriptor: | PATHOGENESIS-RELATED PROTEIN P14A | | Authors: | Fernandez, C, Szyperski, T, Bruyere, T, Ramage, P, Mosinger, E, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the pathogenesis-related protein P14a.
J.Mol.Biol., 266, 1997
|
|
